6G21
 
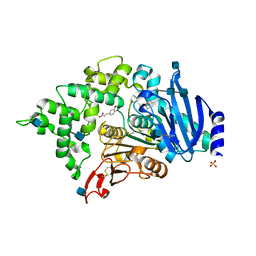 | | Crystal structure of an esterase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an esterase from Aspergillus oryzae
To Be Published
|
|
2CJE
 
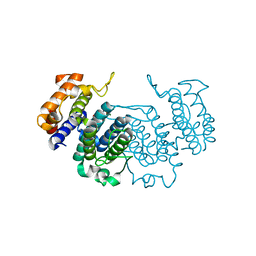 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF Leishmania major DUTPASE WITH SUBSTRATE ANALOGUE DUPNHP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DUTPASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Fogg, M.J, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
6F1J
 
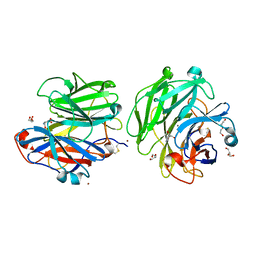 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2BTU
 
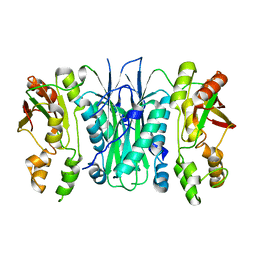 | | Crystal structure of Phosphoribosylformylglycinamidine cyclo-ligase from Bacillus Anthracis at 2.3A resolution. | | Descriptor: | PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE | | Authors: | Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Lebedev, A.A, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-06-07 | | Release date: | 2006-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Phosphoribosylformylglycinamidine Cyclo-Ligase from Bacillus Anthracis at 2.3A Resolution.
To be Published
|
|
2CIC
 
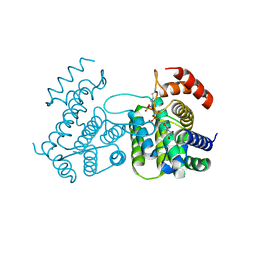 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
2C40
 
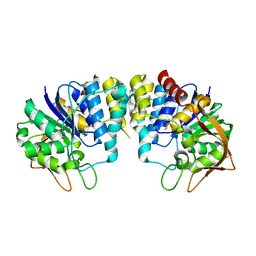 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | Descriptor: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | Authors: | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
6FHJ
 
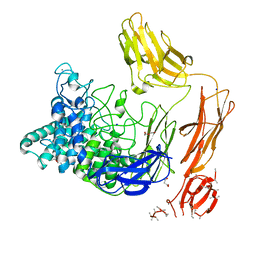 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
4ARO
 
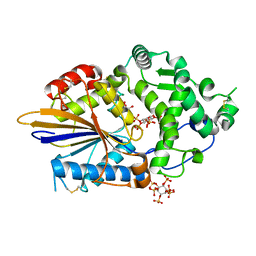 | | Hafnia Alvei phytase in complex with myo-inositol hexakis sulphate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, DI(HYDROXYETHYL)ETHER, HISTIDINE ACID PHOSPHATASE, ... | | Authors: | Moroz, O.V, Blagova, E.B, Ariza, A, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
3ZKW
 
 | |
9F5H
 
 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
6Y5T
 
 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
8QF8
 
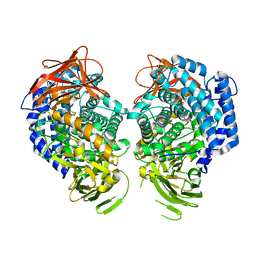 | | GH146 beta-L-arabinofuranosidase from Bacteroides thetaioatomicron in complex with beta-l-arabinofurano cyclophellitol aziridine | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, (1~{S},2~{S},3~{S},4~{S},5~{S})-4-(hydroxymethyl)-6-azabicyclo[3.1.0]hexane-2,3-diol, Glycosyl hydrolase, ... | | Authors: | Borlandelli, V, Offen, W, Moroz, O.V, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-04 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
5TCY
 
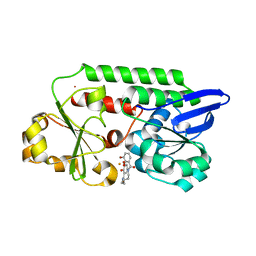 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6HPF
 
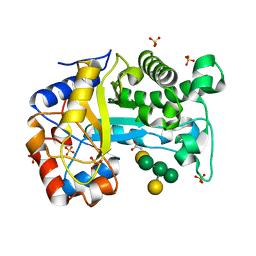 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
1DTO
 
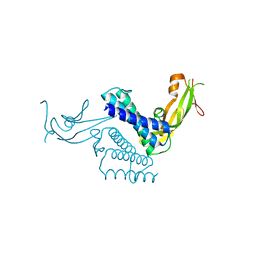 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
8QF2
 
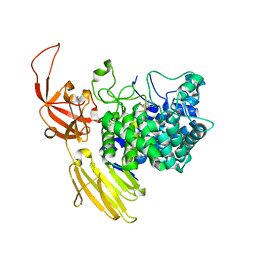 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
6Q7J
 
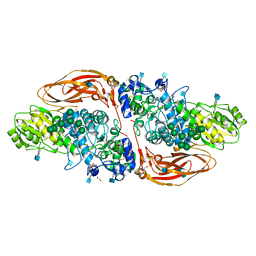 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q7I
 
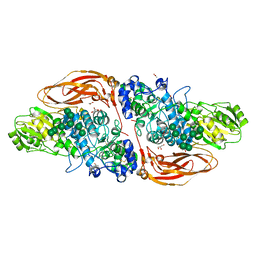 | | GH3 exo-beta-xylosidase (XlnD) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8N
 
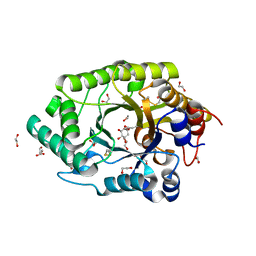 | | GH10 endo-xylanase in complex with xylobiose epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{S},5~{R})-cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8M
 
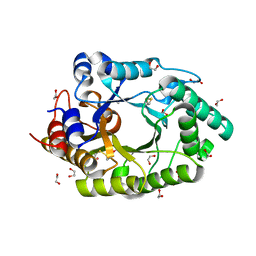 | | GH10 endo-xylanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6SAV
 
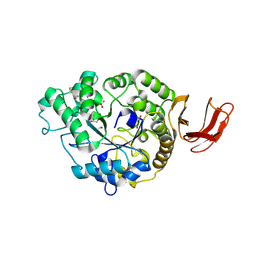 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
2YAZ
 
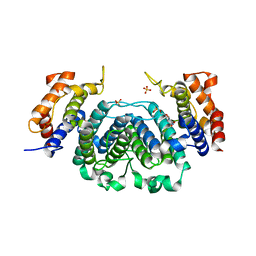 | | The Crystal Structure of Leishmania major dUTPase in complex dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
6SAO
 
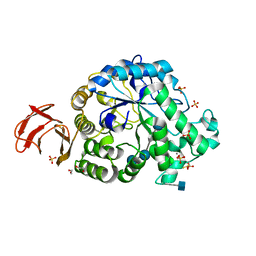 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAU
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, SODIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tinaqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
