4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6CUX
 
 | |
6CUU
 
 | | Thermus thermophiles RNA polymerase in complex with promoter DNA and antibiotic Kanglemycin A | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*CP*TP*CP*TP*GP*AP*TP*GP*CP*A)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*CP*AP*GP*AP*GP*CP*CP*CP*AP*AP*AP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Mode of Action of Kanglemycin A, an Ansamycin Natural Product that Is Active against Rifampicin-Resistant Mycobacterium tuberculosis.
Mol. Cell, 72, 2018
|
|
6DTA
 
 | | Bacteriophage N4 RNA polymerase II elongation complex 2 | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
6DT8
 
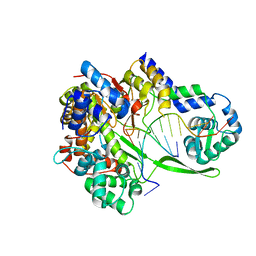 | | Bacteriophage N4 RNA polymerase II elongation complex 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*GP*GP*UP*GP*G)-3'), RNAP1, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
6DT7
 
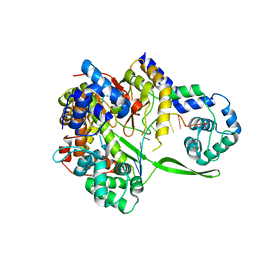 | | Bacteriophage N4 RNA polymerase II and DNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*CP*TP*GP*CP*A)-3'), RNAP1, RNAP2 | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
5UAH
 
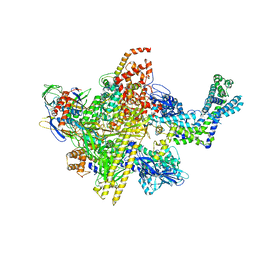 | | Escherichia coli RNA polymerase and Rifampin complex, RpoB D516V mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5UAC
 
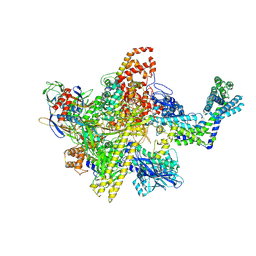 | | Escherichia coli RNA polymerase and Rifampin complex, wild-type | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5UAL
 
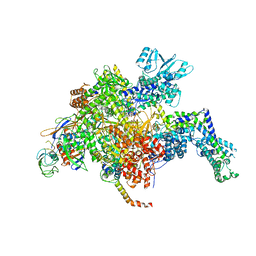 | | Escherichia coli RNA polymerase and Rifampin complex, RpoB S531L mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.887 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6X7K
 
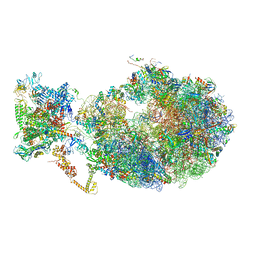 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XIJ
 
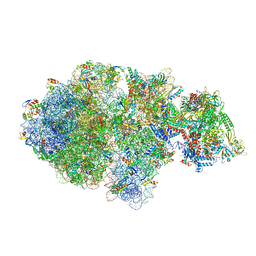 | | Escherichia coli transcription-translation complex A (TTC-A) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
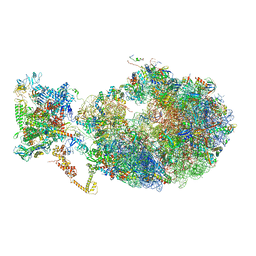 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XGF
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X9Q
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5UAG
 
 | | Escherichia coli RNA polymerase mutant - RpoB D516V | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.399 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6X6T
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B1 (TTC-B1) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5UAQ
 
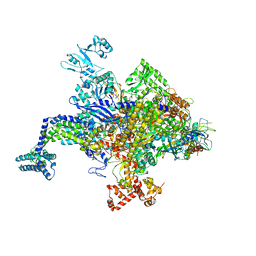 | | Escherichia coli RNA polymerase RpoB H526Y mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5UAJ
 
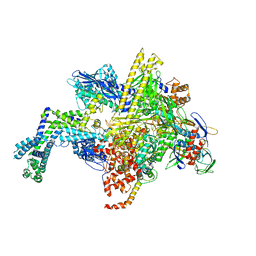 | | Escherichia coli RNA polymerase RpoB S531L mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.915 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6TYF
 
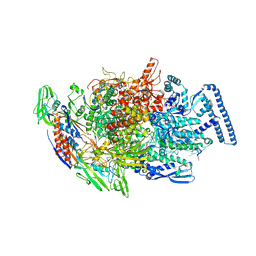 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYG
 
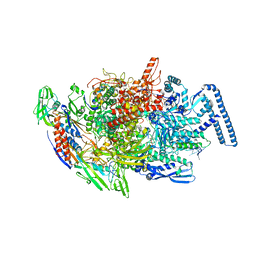 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYE
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VZ5
 
 | | Escherichia coli transcription-translation complex D3 (TTC-D3) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
