6QBA
 
 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with non-retinoid ligand A1120 and engineered binding scaffold | | Descriptor: | 2-[({4-[2-(trifluoromethyl)phenyl]piperidin-1-yl}carbonyl)amino]benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mlynek, G, Brey, C.U, Djinovic-Carugo, K, Puehringer, D. | | Deposit date: | 2018-12-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformation-specific ON-switch for controlling CAR T cells with an orally available drug.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QPI
 
 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
5O4E
 
 | | Crystal structure of VEGF in complex with heterodimeric Fcab JanusCT6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mlynek, G, Lobner, E, Kubinger, K, Humm, A, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
7P0E
 
 | |
7OUV
 
 | |
7OUU
 
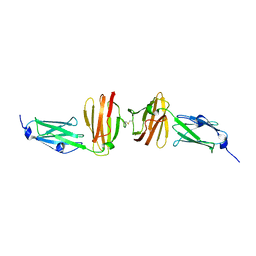 | |
5JII
 
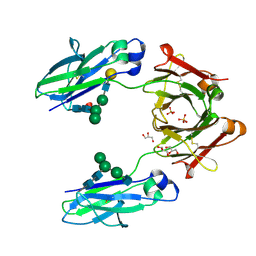 | | Crystal structure of human IgG1-Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
4WWS
 
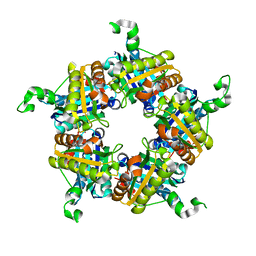 | |
6ERC
 
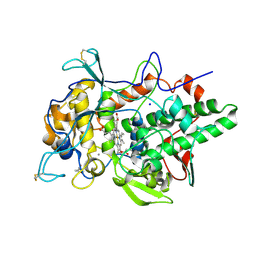 | | Peroxidase A from Dictyostelium discoideum (DdPoxA) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolussi, A, Mlynek, G, Furtmueller, P.G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2017-10-17 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.50001836 Å) | | Cite: | Secreted heme peroxidase from Dictyostelium discoideum: Insights into catalysis, structure, and biological role.
J. Biol. Chem., 293, 2018
|
|
3NN2
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with cyanide | | Descriptor: | CYANIDE ION, Chlorite dismutase, GLYCEROL, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN1
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with imidazole | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN3
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173A mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NN4
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173K mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
5MAU
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7ASB
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
7A8T
 
 | | Crystal structure of sarcomeric protein FATZ-1 (mini-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
7A8U
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
7ANK
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with half dimer of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-10-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
6FKS
 
 | | Crystal structure of a dye-decolorizing peroxidase from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-24 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.60000467 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6FL2
 
 | | Crystal structure of a dye-decolorizing peroxidase D143A variant from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.270001 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
6FIY
 
 | | Crystal structure of a dye-decolorizing peroxidase D143AR232A variant from Klebsiella pneumoniae (KpDyP) | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Hofbauer, S, Mlynek, G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09000432 Å) | | Cite: | Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage.
J. Biol. Chem., 293, 2018
|
|
