1KSO
 
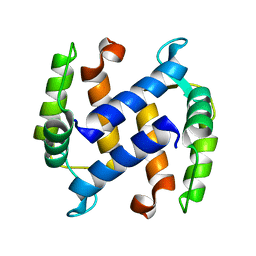 | | CRYSTAL STRUCTURE OF APO S100A3 | | Descriptor: | S100 CALCIUM-BINDING PROTEIN A3 | | Authors: | Mittl, P.R, Fritz, G, Sargent, D.F, Richmond, T.J, Heizmann, C.W, Grutter, M.G. | | Deposit date: | 2002-01-14 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-free MIRAS phasing: structure of apo-S100A3.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4PLQ
 
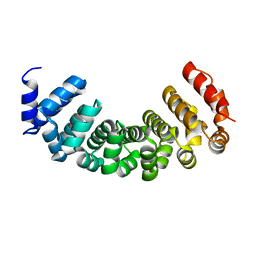 | |
7Z7C
 
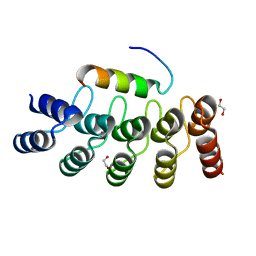 | |
1OUV
 
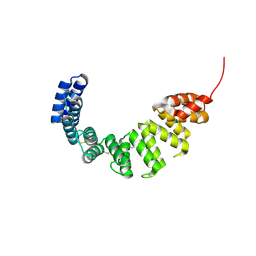 | | Helicobacter cysteine rich protein C (HcpC) | | Descriptor: | conserved hypothetical secreted protein | | Authors: | Mittl, P.R, Luethy, L. | | Deposit date: | 2003-03-25 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Helicobacter Cysteine-rich Protein C at 2.0A Resolution: Similar Peptide-binding Sites in TPR and SEL1-like Repeat Proteins
J.Mol.Biol., 340, 2004
|
|
9F23
 
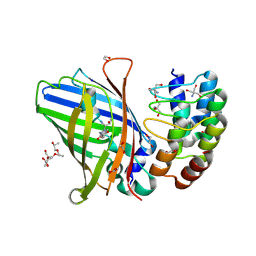 | | DARPin eGFP complex DP2 (2G156) | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mittl, P.R, Hansen, S. | | Deposit date: | 2024-04-22 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | DARPin eGFP complex DP2 (2G156)
To Be Published
|
|
9F22
 
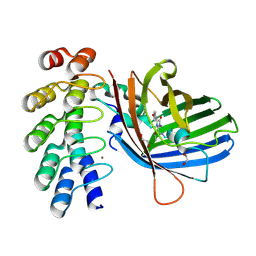 | |
9F24
 
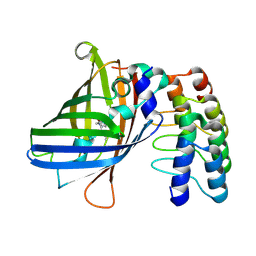 | |
2AXI
 
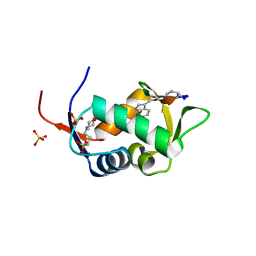 | | HDM2 in complex with a beta-hairpin | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, ... | | Authors: | Mittl, P.R.E, Fasan, R, Robinson, J, Gruetter, M.G. | | Deposit date: | 2005-09-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Studies in a Family of beta-Hairpin Protein Epitope Mimetic Inhibitors of the p53-HDM2 Protein-Protein Interaction.
Chembiochem, 7, 2006
|
|
1TGK
 
 | |
1TGJ
 
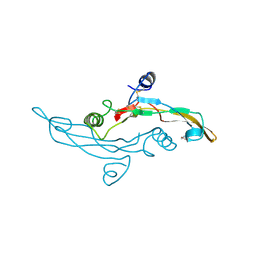 | | HUMAN TRANSFORMING GROWTH FACTOR-BETA 3, CRYSTALLIZED FROM DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, TRANSFORMING GROWTH FACTOR-BETA 3 | | Authors: | Mittl, P.R.E, Priestle, J.P, Gruetter, M.G. | | Deposit date: | 1996-07-09 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding.
Protein Sci., 5, 1996
|
|
8A1A
 
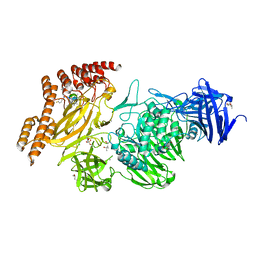 | | Structure of a leucinostatin derivative determined by host lattice display : L1F11V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8A19
 
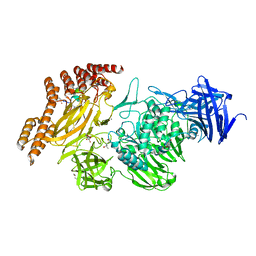 | | Structure of a leucinostatin derivative determined by host lattice display : L1E4V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1AIE
 
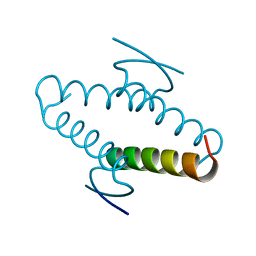 | |
7BHE
 
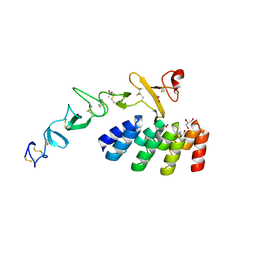 | | DARPin_D5/Her3 domain 4 complex, monoclinic crystals | | Descriptor: | ACETATE ION, DARPin_D5, GLYCEROL, ... | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7BHF
 
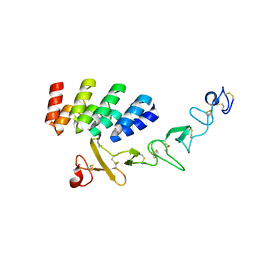 | | DARPin_D5/Her3 domain 4 complex, orthorhombic crystals | | Descriptor: | ACETATE ION, DARPin_D5, Isoform 4 of Receptor tyrosine-protein kinase erbB-3 | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1GES
 
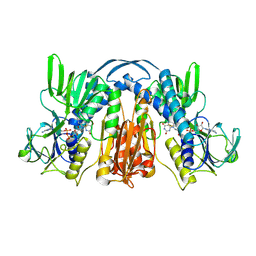 | |
1GEU
 
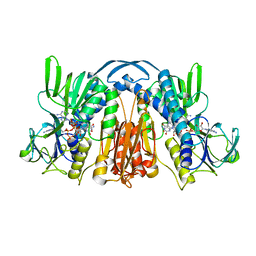 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GET
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2DKO
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | Caspase-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Mittl, P.R.E, Ganesan, R, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
1CP3
 
 | |
1GER
 
 | |
4ATZ
 
 | |
5OD1
 
 | | Structure of the engineered metalloesterase MID1sc10 complexed with a phosphonate transition state analogue | | Descriptor: | GLYCEROL, MID1sc10, ZINC ION, ... | | Authors: | Mittl, P.R.E, Studer, S, Hansen, D.A, Hilvert, D. | | Deposit date: | 2017-07-04 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
6F5E
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, DD_D12_10_47, Mitogen-activated protein kinase 8 | | Authors: | Wu, Y, Mittl, P.R, Honegger, A, Batyuk, A, Plueckthun, A. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
To be published
|
|
