4U12
 
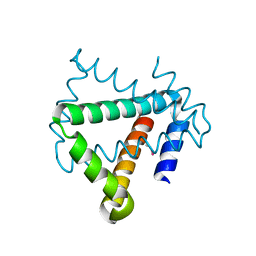 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | Descriptor: | Uncharacterized protein HP0242 | | Authors: | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
4U13
 
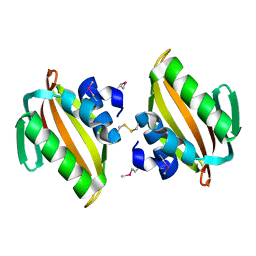 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | Descriptor: | putative polyketide cyclase SMa1630 | | Authors: | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-14 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
7Q18
 
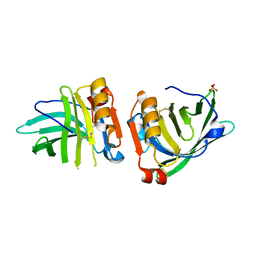 | | Beta-lactoglobulin mutant FAF (I56F/L39A/M107F), unliganded form | | Descriptor: | Beta-lactoglobulin, SULFATE ION | | Authors: | Loch, J.I, Cymborowski, M.T, Minor, W, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
6EDD
 
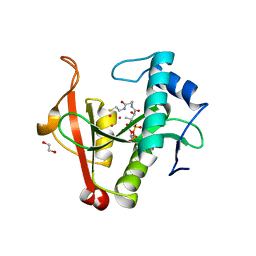 | | Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6EDV
 
 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
5N0H
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5NBK
 
 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N0I
 
 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
6WUW
 
 | | Crystal structure of Human Serum Albumin complex with JMS-053 | | Descriptor: | 1,2-ETHANEDIOL, 7-imino-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione, MYRISTIC ACID, ... | | Authors: | Czub, M.P, Cooper, D.R, Shabalin, I.G, Lazo, J.S, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Complex of an Iminopyridinedione Protein Tyrosine Phosphatase 4A3 Phosphatase Inhibitor with Human Serum Albumin.
Mol.Pharmacol., 98, 2020
|
|
6XK0
 
 | | Albumin-dexamethasone complex | | Descriptor: | Albumin, CITRATE ANION, DEXAMETHASONE, ... | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2020-06-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Iucrj, 7, 2020
|
|
6S0S
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0W
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0T
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | Descriptor: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0U
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0V
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, neamine and sulfate | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0R
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | Descriptor: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6E5Z
 
 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5END
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(Q152A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL | | Authors: | Hou, J, Cooper, D.R, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
5EPE
 
 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
6M8Z
 
 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6MDQ
 
 | | Crystal structure of equine serum albumin in complex with testosterone | | Descriptor: | CITRATE ANION, Serum albumin, TESTOSTERONE | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Handing, K.B, Venkataramany, B.S, Cymborowski, M.T, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-05 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Testosterone meets albumin - the molecular mechanism of sex hormone transport by serum albumins.
Chem Sci, 10, 2019
|
|
5HOZ
 
 | |
5IIU
 
 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
