2QGO
 
 | |
3PAN
 
 | |
2PBZ
 
 | |
2XAH
 
 | | Crystal structure of LSD1-CoREST in complex with (+)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | 3-PHENYLPROPANAL, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAS
 
 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2580, 14e) | | Descriptor: | 3-[4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)PHENYL]PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
4KD6
 
 | | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M
To be Published
|
|
2OTE
 
 | | Crystal structure of a monomeric cyan fluorescent protein in the photobleached state | | Descriptor: | ACETATE ION, GFP-like fluorescent chromoprotein cFP484 | | Authors: | Henderson, J.N, Ai, H, Campbell, R.E, Remington, S.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for reversible photobleaching of a green fluorescent protein homologue.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OTB
 
 | |
2HXP
 
 | | Crystal Structure of the human phosphatase (DUSP9) | | Descriptor: | Dual specificity protein phosphatase 9, PHOSPHATE ION | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3OR5
 
 | |
3OXN
 
 | |
4M1V
 
 | | Crystal structure of the ancestral soluble variant of the Human Phosphate Binding Protein (HPBP) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Gonzalez, D, Hiblot, J, Darbinian, N, Miller, J.S, Gotthard, G, Amini, S, Chabriere, E, Elias, M. | | Deposit date: | 2013-08-04 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ancestral mutations as a tool for solubilizing proteins: The case of a hydrophobic phosphate-binding protein.
FEBS Open Bio, 4, 2014
|
|
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3SC2
 
 | | REFINED ATOMIC MODEL OF WHEAT SERINE CARBOXYPEPTIDASE II AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE II (CPDW-II), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liao, D.-I, Remington, S.J. | | Deposit date: | 1992-07-01 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined atomic model of wheat serine carboxypeptidase II at 2.2-A resolution.
Biochemistry, 31, 1992
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
3KTO
 
 | |
4O8E
 
 | | Crystal structure of the complex of type I ribosome inactivating protein from Momordica balsamina with uridine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Pandey, S, Yamini, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein from Momordica balsamina with uridine triphosphate at 2.0 A resolution
To be Published
|
|
4UVA
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
3KZG
 
 | |
6F8E
 
 | | PH domain from TgAPH | | Descriptor: | Pleckstrin homology domain | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Phosphatidic Acid Sensing by APH in Apicomplexan Parasites.
Structure, 26, 2018
|
|
3LJS
 
 | |
3LDT
 
 | |
2LZK
 
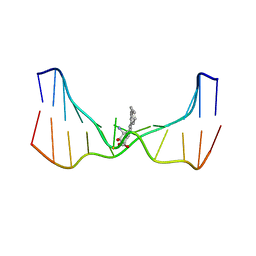 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
4J3C
 
 | | Crystal structure of 16S ribosomal RNA methyltransferase RsmE | | Descriptor: | Ribosomal RNA small subunit methyltransferase E | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 16S ribosomal RNA methyltransferase RsmE
TO BE PUBLISHED
|
|
