3BX2
 
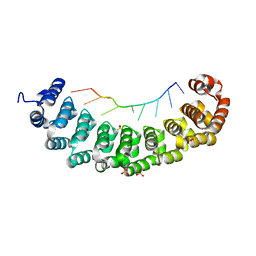 | | Puf4 RNA binding domain bound to HO endonuclease RNA 3' UTR recognition sequence | | Descriptor: | HO endonuclease 3' UTR binding sequence, Protein PUF4, SODIUM ION, ... | | Authors: | Miller, M.T, Higgin, J.J, Hall, T.M.T. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p
Nat.Struct.Mol.Biol., 15, 2008
|
|
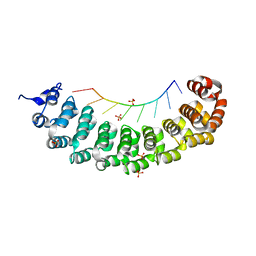
3BX3
 
 | |
3BWT
 
 | |
5D3G
 
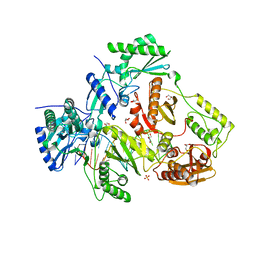 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
3POY
 
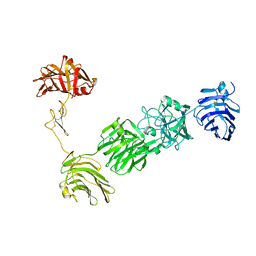 | | Crystal Structure of the alpha-Neurexin-1 ectodomain, LNS 2-6 | | Descriptor: | Neurexin-1-alpha, beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Miller, M.T, Mileni, M, Comoletti, D, Stevens, R.C, Harel, M, Taylor, P. | | Deposit date: | 2010-11-23 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The crystal structure of the alpha-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function.
Structure, 19, 2011
|
|
1JGT
 
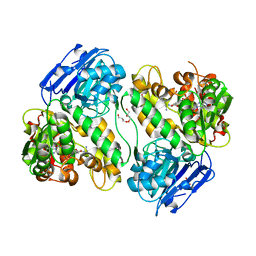 | | CRYSTAL STRUCTURE OF BETA-LACTAM SYNTHETASE | | Descriptor: | BETA-LACTAM SYNTHETASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GLYCEROL, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of beta-lactam synthetase reveals how to synthesize antibiotics instead of asparagine.
Nat.Struct.Biol., 8, 2001
|
|
1M1Z
 
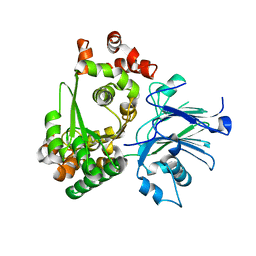 | |
1MC1
 
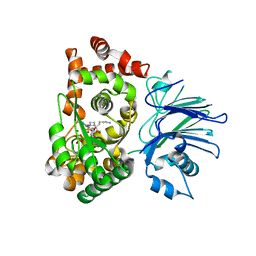 | | BETA-LACTAM SYNTHETASE WITH PRODUCT (DGPC), AMP AND PPI | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-LACTAM SYNTHETASE, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MB9
 
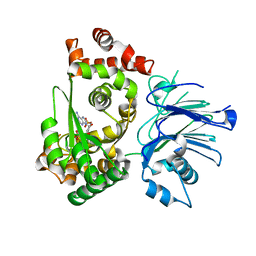 | | BETA-LACTAM SYNTHETASE COMPLEXED WITH ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, BETA-LACTAM SYNTHETASE, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-02 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MBZ
 
 | | BETA-LACTAM SYNTHETASE WITH TRAPPED INTERMEDIATE | | Descriptor: | ARGININE-N-METHYLCARBONYL PHOSPHORIC ACID 5'-ADENOSINE ESTER, BETA-LACTAM SYNTHETASE, GLYCEROL, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q19
 
 | | Carbapenam Synthetase | | Descriptor: | (2S,5S)-5-CARBOXYMETHYLPROLINE, CarA, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
1Q15
 
 | | Carbapenam Synthetase | | Descriptor: | CarA | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
4WEB
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMAMIDE, Mouse Fab Heavy Chain, ... | | Authors: | Khan, A.G, Whidby, J, Miller, M.T, Scarborough, H, Zatorski, A.V, Cygan, A, Price, A.A, Yost, S.A, Bohannon, C.D, Jacob, J, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2.
Nature, 509, 2014
|
|
5F9F
 
 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3BE8
 
 | | Crystal structure of the synaptic protein neuroligin 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-01-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and Its beta-Neurexin Complex: Determinants for Folding and Cell Adhesion
Neuron, 56, 2007
|
|
4P7H
 
 | |
3NHZ
 
 | | Structure of N-terminal Domain of MtrA | | Descriptor: | MAGNESIUM ION, Two component system transcriptional regulator mtrA | | Authors: | Barbieri, C.M, Mack, T.R, Robinson, V.L, Miller, M.T, Stock, A.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
4PA0
 
 | | Omecamtiv Mercarbil binding site on the Human Beta-Cardiac Myosin Motor Domain | | Descriptor: | GLYCEROL, Myosin-7,Green fluorescent protein, methyl 4-(2-fluoro-3-{[(6-methylpyridin-3-yl)carbamoyl]amino}benzyl)piperazine-1-carboxylate | | Authors: | Winkelmann, D.A, Miller, M.T, Stock, A.M. | | Deposit date: | 2014-04-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity.
Nat Commun, 6, 2015
|
|
5E3H
 
 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
2WQZ
 
 | | Crystal structure of synaptic protein neuroligin-4 in complex with neurexin-beta 1: alternative refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEUREXIN-1-BETA, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and its Beta-Neurexin Complex: Determinants for Folding and Cell Adhesion.
Neuron, 56, 2007
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
