8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FPE
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5D69
 
 | | Human calpain PEF(S) with (2Z,2Z')-2,2'-disulfanediylbis(3-(6-iodoindol-3-yl)acrylic acid) bound | | Descriptor: | (2E,2'Z)-2,2'-disulfanediylbis[3-(4-iodophenyl)prop-2-enoic acid], CALCIUM ION, Calpain small subunit 1, ... | | Authors: | Adams, S.E, Robinson, E.J, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
2AQ7
 
 | | Structure-activity relationships at the 5-posiiton of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherichia coli | | Descriptor: | (5R)-4-HYDROXY-3,5-DIMETHYL-5-[(1E,3E)-2-METHYLPENTA-1,3-DIENYL]THIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
2AQB
 
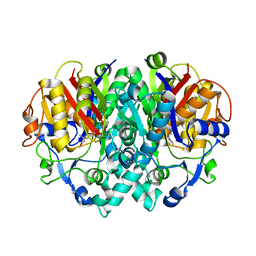 | | Structure-activity relationships at the 5-position of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherchia coli | | Descriptor: | (5R)-5-[(1E)-BUTA-1,3-DIENYL]-4-HYDROXY-3,5-DIMETHYLTHIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
4PHN
 
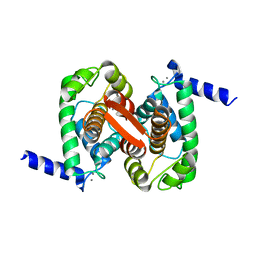 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Allemann, R.K, Rizkallah, P.J, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4PHJ
 
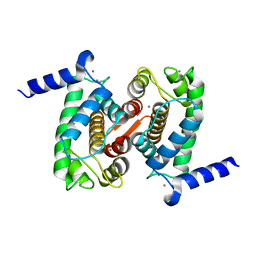 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids: Human unliganded protein | | Descriptor: | CALCIUM ION, Calpain small subunit 1 | | Authors: | Adams, S.E, Rizkallah, P.J, Allemann, R.K, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4PHM
 
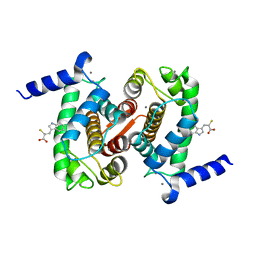 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids | | Descriptor: | 3-(5-bromo-1H-indol-3-yl)-2-thioxopropanoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4PHK
 
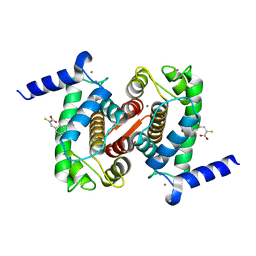 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids. The complex with (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid | | Descriptor: | (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
4RG9
 
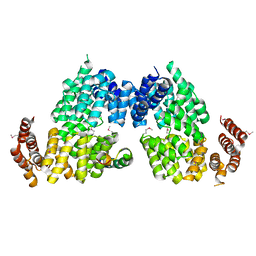 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4RG7
 
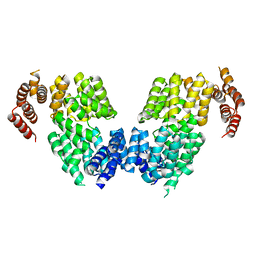 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4R32
 
 | | Crystal Structure Analysis of Pyk2 and Paxillin LD motifs | | Descriptor: | Paxillin, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.D, Nourse, A, Zheng, J.J. | | Deposit date: | 2014-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
4RG6
 
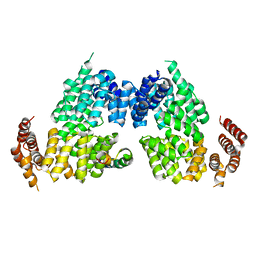 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
8E3N
 
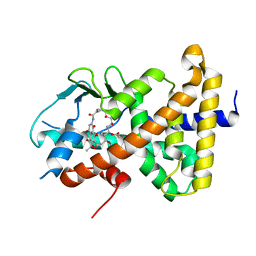 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5UTO
 
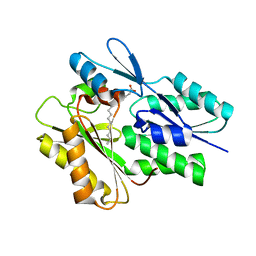 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, Broussard, T.C, Miller, D.J, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-15 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
8F5Y
 
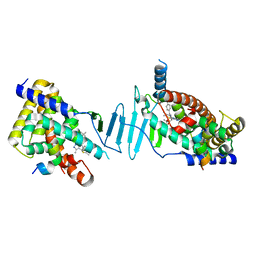 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
3IVQ
 
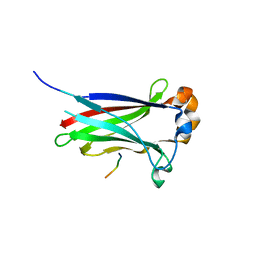 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
3CKE
 
 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
3G4D
 
 | | Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis | | Descriptor: | (+)-delta-cadinene synthase isozyme XC1, BETA-MERCAPTOETHANOL, GLYCEROL | | Authors: | Gennadios, H.A, Di Costanzo, L, Miller, D.J, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
7JLS
 
 | | RV3666c bound to tripeptide | | Descriptor: | Peptide SER-VAL-ALA, Probable periplasmic dipeptide-binding lipoprotein DppA | | Authors: | Lee, R.E, Griffith, E.C, Miller, D.J. | | Deposit date: | 2020-07-30 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Biophysical analysis of the Mycobacteria tuberculosis peptide binding protein DppA reveals a stringent peptide binding pocket.
Tuberculosis (Edinb), 132, 2021
|
|
