5LI1
 
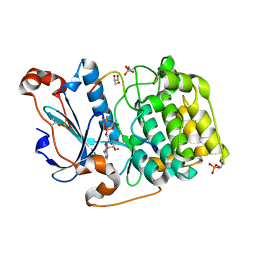 | | Structure of a Par3-inhibitory peptide bound to PKCiota core kinase domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
2MC3
 
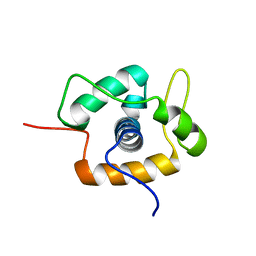 | |
5AMN
 
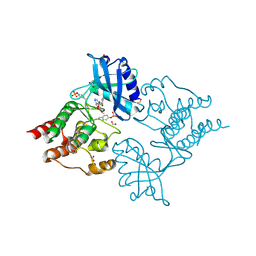 | | The Discovery of 2-Substituted Phenol Quinazolines as Potent and Selective RET Kinase Inhibitors | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Newton, R, Bowler, K, Burns, E.M, Chapman, P, Fairweather, E, Fritzl, S, Goldberg, K, Hamilton, N.M, Holt, S.V, Hopkins, G.V, Jones, S.D, Jordan, A.M, Lyons, A, McDonald, N.Q, Maguire, L.A, Mould, D.P, Purkiss, A.G, Small, H.F, Stowell, A, Thomson, G.J, Waddell, I.D, Waszkowycz, B, Watson, A.J, Ogilvie, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The discovery of 2-substituted phenol quinazolines as potent RET kinase inhibitors with improved KDR selectivity.
Eur J Med Chem, 112, 2016
|
|
1BUN
 
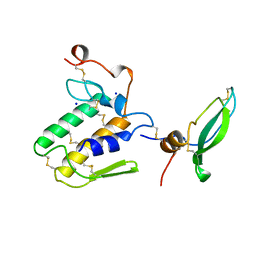 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1E41
 
 | |
1E3Y
 
 | |
1F5Q
 
 | | CRYSTAL STRUCTURE OF MURINE GAMMA HERPESVIRUS CYCLIN COMPLEXED TO HUMAN CYCLIN DEPENDENT KINASE 2 | | Descriptor: | CHLORIDE ION, CYCLIN DEPENDENT KINASE 2, GAMMA HERPESVIRUS CYCLIN | | Authors: | Card, G.L, Knowles, P, Laman, H, Jones, N, McDonald, N.Q. | | Deposit date: | 2000-06-15 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a gamma-herpesvirus cyclin-cdk complex.
EMBO J., 19, 2000
|
|
1GR0
 
 | | myo-inositol 1-phosphate synthase from Mycobacterium tuberculosis in complex with NAD and zinc. | | Descriptor: | CACODYLATE ION, INOSITOL-3-PHOSPHATE SYNTHASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Norman, R.A, Murray-Rust, J, McDonald, N.Q, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Structure, 10, 2002
|
|
2X2M
 
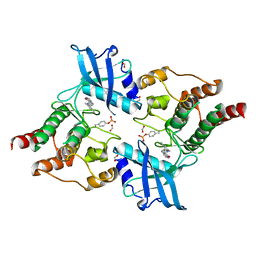 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-3-[(3,5-DIMETHYL-1H-PYRROL-2-YL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
2XQN
 
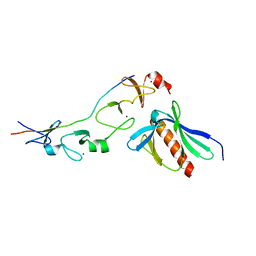 | | Complex of the 2nd and 3rd LIM domains of TES with the EVH1 DOMAIN of MENA and the N-Terminal domain of actin-like protein Arp7A | | Descriptor: | ACTIN-LIKE PROTEIN 7A, ENABLED HOMOLOG, TESTIN, ... | | Authors: | Knowles, P.P, Briggs, D.C, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2010-09-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
J. Biol. Chem., 286, 2011
|
|
2YJE
 
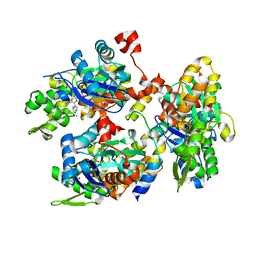 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
2YJF
 
 | | Oligomeric assembly of actin bound to MRTF-A | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Langer, C.A, Guettler, S, McDonald, N.Q, Treisman, R. | | Deposit date: | 2011-05-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator.
Sci Signal, 4, 2011
|
|
4B1X
 
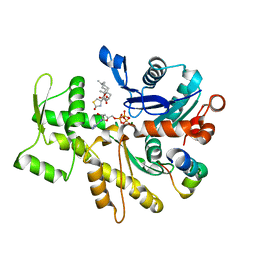 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4CKI
 
 | | Crystal Structure of oncogenic RET tyrosine kinase M918T bound to adenosine | | Descriptor: | ADENOSINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Goodman, K, Martinez-Torres, R.J, Borg, A, Murray-Rust, J, Mouilleron, S, Knowles, P, McDonald, N.Q. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans.
Mol. Cell, 53, 2014
|
|
2J16
 
 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2IVS
 
 | | Crystal structure of non-phosphorylated RET tyrosine kinase domain | | Descriptor: | 2',3'- cyclic AMP, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Chemical Inhibition of the Ret Tyrosine Kinase Domain.
J.Biol.Chem., 281, 2006
|
|
2IVU
 
 | |
2J17
 
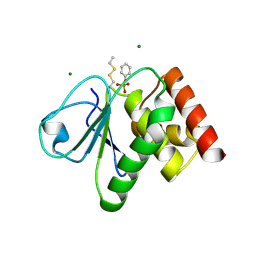 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2IVT
 
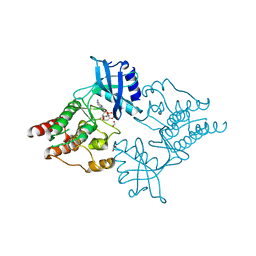 | |
2IVV
 
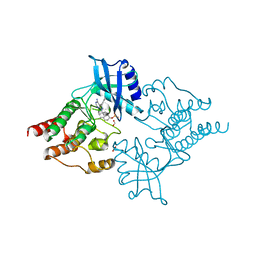 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
2IYB
 
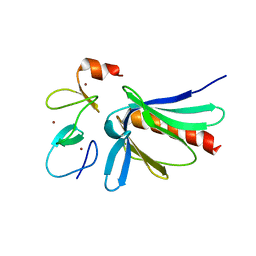 | |
4B1Z
 
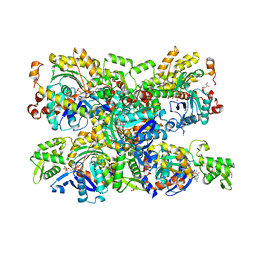 | | Structure of the Phactr1 RPEL domain bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4BG6
 
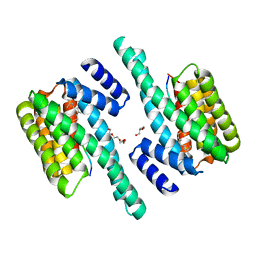 | | 14-3-3 interaction with Rnd3 prenyl-phosphorylation motif | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FARNESYL, ... | | Authors: | Riou, P, Kjaer, S, Purkiss, A, O'Reilly, N, McDonald, N.Q. | | Deposit date: | 2013-03-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 Proteins Interact with a Hybrid Prenyl-Phosphorylation Motif to Inhibit G Proteins.
Cell(Cambridge,Mass.), 153, 2013
|
|
4B1W
 
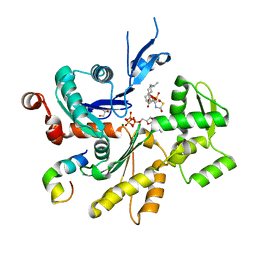 | | Structure of the Phactr1 RPEL-2 domain bound to actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1Y
 
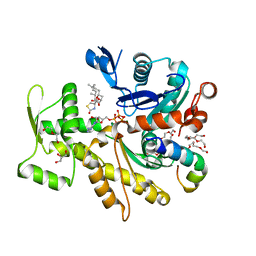 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
