2ATE
 
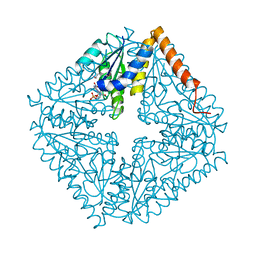 | | Structure of the complex of PurE with NitroAIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kappock, T.J, Mathews, I.I, Zaugg, J.B, Peng, P, Hoskins, A.A, Okamoto, A, Ealick, S.E, Stubbe, J. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
Biochemistry, 46, 2007
|
|
8DE1
 
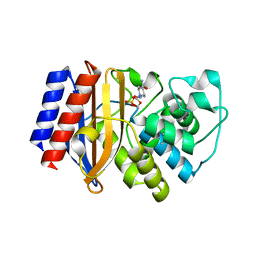 | |
8DE2
 
 | | TEM-1 beta-lactamase A237Y mutant covalently bound to avibactam, a room temperature structure | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DE0
 
 | | TEM-1 beta-lactamase covalently bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis for LAG-3 dimeric association and inhibition of T cell function
To Be Published
|
|
8EE1
 
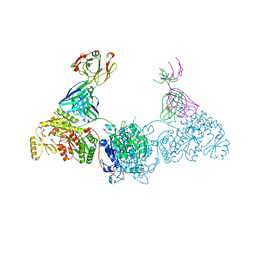 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment AA5 | | Descriptor: | 6-deoxyerythronolide B synthase, AA5 antibody heavy chain, AA5 antibody light chain | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
8ECU
 
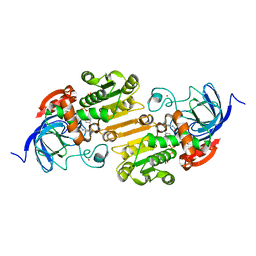 | |
8ECS
 
 | |
8ECT
 
 | |
8EIW
 
 | | Cobalt(II)-substituted Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8EIY
 
 | | Cobalt(II)-substituted S48T Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8EIX
 
 | | Cobalt(II)-substituted S48A Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8DDZ
 
 | | TEM-1 beta-lactamase A237Y | | Descriptor: | Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8EE0
 
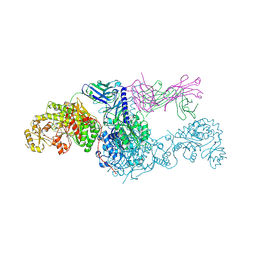 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment 1B2 | | Descriptor: | 1B2 antibody heavy chain, 1B2 antibody light chain, 6-deoxyerythronolide B synthase | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
1YR2
 
 | |
2NM0
 
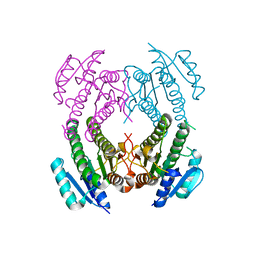 | | Crystal Structure of SCO1815: a Beta-Ketoacyl-Acyl Carrier Protein Reductase from Streptomyces coelicolor A3(2) | | Descriptor: | Probable 3-oxacyl-(Acyl-carrier-protein) reductase | | Authors: | Khosla, C, Tang, Y, Lee, H.Y, Tang, Y, Kim, C.Y, Mathews, I.I. | | Deposit date: | 2006-10-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2).
Biochemistry, 45, 2006
|
|
3RGA
 
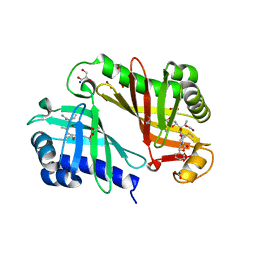 | | Crystal structure of epoxide hydrolase for polyether lasalocid A biosynthesis | | Descriptor: | (4R,5S)-3-[(2R)-2-{(2S,2'R,4S,5S,5'R)-2,5'-diethyl-5'-[(1S)-1-hydroxyethyl]-4-methyloctahydro-2,2'-bifuran-5-yl}butanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, (4R,5S)-3-[(2R,3S,4S)-2-ethyl-5-[(3R)-2-ethyl-3-[2-[(2R,3R)-2-ethyl-3-methyl-oxiran-2-yl]ethyl]oxiran-2-yl]-3-hydroxy-4-methyl-pentanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, ACETATE ION, ... | | Authors: | Hotta, K, Mathews, I.I, Chen, X, Kim, C.-Y. | | Deposit date: | 2011-04-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enzymatic catalysis of anti-Baldwin ring closure in polyether biosynthesis
Nature, 483, 2012
|
|
3SL2
 
 | | ATP Forms a Stable Complex with the Essential Histidine Kinase WalK (YycG) Domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Sensor histidine kinase yycG | | Authors: | Celikel, R, Veldore, V.H, Mathews, I, Devine, K, Varughese, K.I. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | ATP forms a stable complex with the essential histidine kinase WalK (YycG) domain.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RGI
 
 | |
1NRS
 
 | |
3SBM
 
 | | Trans-acting transferase from Disorazole synthase in complex with Acetate | | Descriptor: | ACETATE ION, DisD protein, HEXAETHYLENE GLYCOL | | Authors: | Khosla, C, Mathews, I.I, Wong, F.T, Jin, X. | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Mechanism of the trans-Acting Acyltransferase from the Disorazole Synthase.
Biochemistry, 50, 2011
|
|
1C3Q
 
 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
7MHA
 
 | |
1ESQ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
7LAV
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
