7WD3
 
 | | Cryo-EM structure of Drg1 hexamer treated with ATP and benzo-diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WBB
 
 | | Cryo-EM structure of substrate engaged Drg1 hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AFG2 isoform 1, substrate | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
5OPI
 
 | | Crystal structure of the TAPBPR-MHC I peptide editing complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Thomas, C, Tampe, R. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing.
Science, 358, 2017
|
|
7YKT
 
 | | Cryo-EM structure of Drg1 hexamer in helical state treated with ADP/AMPPNP/benzo-diazaborine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKZ
 
 | | Cryo-EM structure of Drg1 hexamer in the planar state treated with ADP/AMPPNP/Diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKL
 
 | | Cryo-EM structure of Drg1 hexamer treated with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKK
 
 | | Cryo-EM structure of Drg1 hexamer treated with ADP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
8F2P
 
 | |
4DEP
 
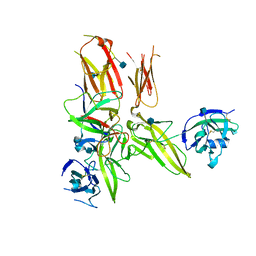 | | Structure of the IL-1b signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, Interleukin-1 receptor accessory protein, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2012-01-21 | | Release date: | 2012-03-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the activating IL-1 receptor signaling complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2WBL
 
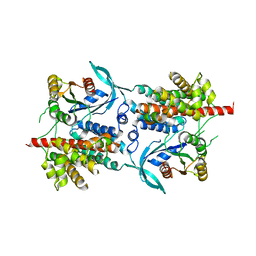 | | Three-dimensional structure of a binary ROP-PRONE complex | | Descriptor: | RAC-LIKE GTP-BINDING PROTEIN ARAC2, RHO OF PLANTS GUANINE NUCLEOTIDE EXCHANGE FACTOR 8 | | Authors: | Thomas, C, Fricke, I, Weyand, M, Berken, A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Biol.Chem., 390, 2009
|
|
1H10
 
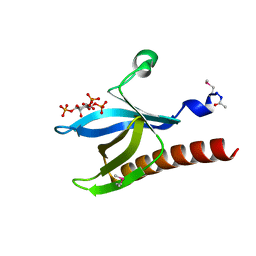 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
6IL3
 
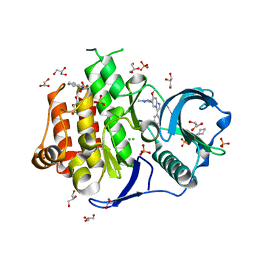 | | Crystal structure of the FLT3 kinase bound to a small molecule inhibitor | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, 7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)-3-(pyridin-2-yl)imidazo[1,2-a]pyridine, CHLORIDE ION, ... | | Authors: | Thomas, C.J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the FLT3 kinase bound to a small molecule inhibitor
To Be Published
|
|
3K4T
 
 | |
1SVS
 
 | | Structure of the K180P mutant of Gi alpha subunit bound to GppNHp. | | Descriptor: | Guanine nucleotide-binding protein G(i), alpha-1 subunit, MAGNESIUM ION, ... | | Authors: | Thomas, C.J, Du, X, Li, P, Wang, Y, Ross, E.M, Sprang, S.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein {alpha}-subunit.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1EAZ
 
 | | Crystal structure of the phosphoinositol (3,4)-bisphosphate binding PH domain of TAPP1 from human. | | Descriptor: | CITRIC ACID, TANDEM PH DOMAIN CONTAINING PROTEIN-1 | | Authors: | Thomas, C.C, Dowler, S, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Phosphatidylinositol 3,4-Bisphosphate-Binding Pleckstrin Homology (Ph) Domain of Tandem Ph-Domain-Containing Protein 1 (Tapp1): Molecular Basis of Lipid Specificity
Biochem.J., 358, 2001
|
|
3SE4
 
 | | human IFNw-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha/beta receptor 1, Interferon alpha/beta receptor 2, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3S98
 
 | | human IFNAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha/beta receptor 1 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3S8W
 
 | | D2 domain of human IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-05-31 | | Release date: | 2011-08-31 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3S9D
 
 | | binary complex between IFNa2 and IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9999 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
1T95
 
 | |
3SE3
 
 | | human IFNa2-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha 2b, Interferon alpha/beta receptor 1, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.0001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
2NTX
 
 | | Prone8 | | Descriptor: | Emb|CAB41934.1 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
1NW5
 
 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to S-ADENOSYLMETHIONINE | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, S-ADENOSYLMETHIONINE | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NW6
 
 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to sinefungin | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, SINEFUNGIN | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
2NTY
 
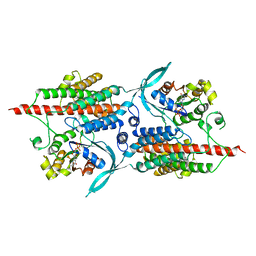 | | Rop4-GDP-PRONE8 | | Descriptor: | Emb|CAB41934.1, GUANOSINE-5'-DIPHOSPHATE, Rac-like GTP-binding protein ARAC5 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
