8J6R
 
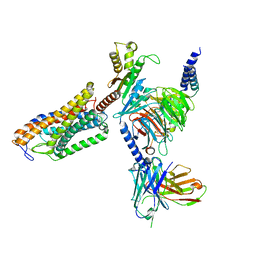 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6P
 
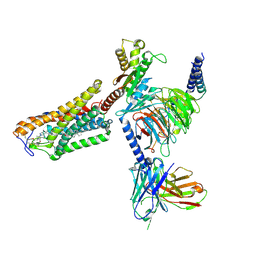 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6Q
 
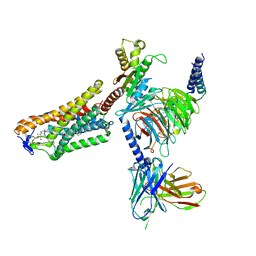 | | Cryo-EM structure of the 3-HB and compound 9n-bound human HCAR2-Gi1 complex | | Descriptor: | (3R)-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8G59
 
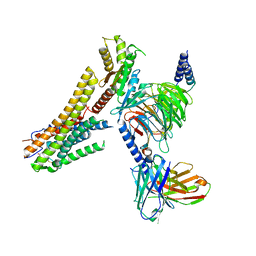 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
1A9T
 
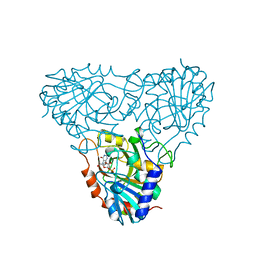 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH 9-DEAZAINOSINE AND PHOSPHATE | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9O
 
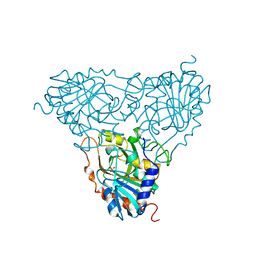 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9S
 
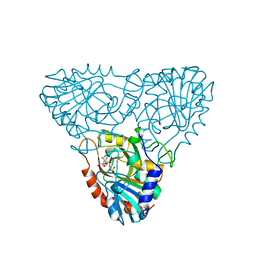 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH INOSINE AND SULFATE | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9R
 
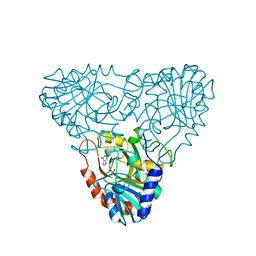 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH HYPOXANTHINE AND SULFATE | | Descriptor: | HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9Q
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH INOSINE | | Descriptor: | HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9P
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH 9-DEAZAINOSINE AND PHOSPHATE | | Descriptor: | 9-DEAZAINOSINE, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1K2P
 
 | | Crystal structure of Bruton's tyrosine kinase domain | | Descriptor: | Tyrosine-protein kinase BTK | | Authors: | Mao, C, Zhou, M, Uckun, F.M. | | Deposit date: | 2001-09-28 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bruton's tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked agammaglobulinemia.
J.Biol.Chem., 276, 2001
|
|
8IKL
 
 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
1ECP
 
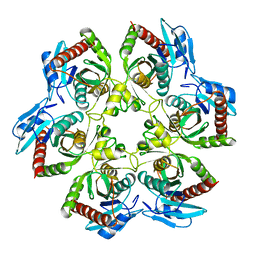 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-13 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
Structure, 5, 1997
|
|
1PBN
 
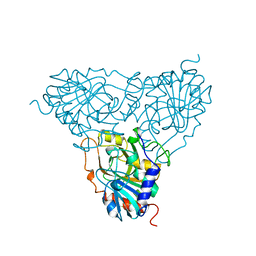 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-10 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
7C7Q
 
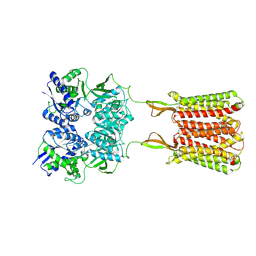 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
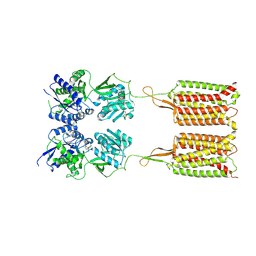 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
6WK7
 
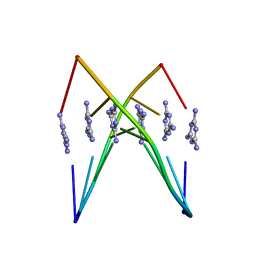 | | Crystal Structure Analysis of a poly(thymine) DNA duplex | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, DNA (5'-D(*TP*TP*TP*TP*TP*T)-3') | | Authors: | Li, Q, Zhao, J, Liu, L, Mandal, S, Rizzuto, F.J, He, H, Wei, S, Jonchhe, S, Sleiman, H.F, Mao, H, Mao, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | A poly(thymine)-melamine duplex for the assembly of DNA nanomaterials.
Nat Mater, 19, 2020
|
|
7R96
 
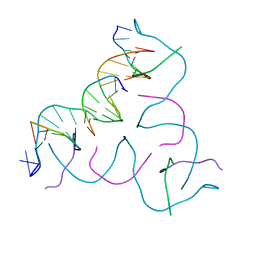 | | Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Sha, R, Mao, C, Seeman, N.C. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (5.68 Å) | | Cite: | 3D Hexagonal Arrangement of DNA Tensegrity Triangles.
Acs Nano, 15, 2021
|
|
8CS8
 
 | | [High G:C, High Vapor Diffusion] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*GP*CP*CP*TP*GP*CP*GP*CP*GP*GP*AP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
