1KY9
 
 | | Crystal Structure of DegP (HtrA) | | Descriptor: | PROTEASE DO | | Authors: | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2002-02-04 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
2WL8
 
 | | X-ray crystal structure of Pex19p | | Descriptor: | PEROXISOMAL BIOGENESIS FACTOR 19 | | Authors: | Schueller, N, Holton, S.J, Stanley, W.A, Song, Y.H, Konarev, P, Roessle, M, Erdmann, R, Schliebs, W, Wilmanns, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Embo J., 29, 2010
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
1JO8
 
 | | Structural analysis of the yeast actin binding protein Abp1 SH3 domain | | Descriptor: | ACTIN BINDING PROTEIN, SULFATE ION | | Authors: | Fazi, B, Cope, M.J, Douangamath, A, Ferracuti, S, Schirwitz, K, Zucconi, A, Drubin, D.G, Wilmanns, M, Cesareni, G, Castagnoli, L. | | Deposit date: | 2001-07-27 | | Release date: | 2002-03-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis.
J.Biol.Chem., 277, 2002
|
|
3H0C
 
 | | Crystal Structure of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-({(2S)-1-[(3R)-3-amino-4-(3-chlorophenyl)butanoyl]pyrrolidin-2-yl}methyl)-3-(methylsulfonyl)benzamide | | Authors: | Nordhoff, S, Cerezo-Galvez, S, Deppe, H, Hill, O, Lopez-Canet, M, Rummey, C, Thiemann, M, Matassa, V.G, Edwards, P.J, Feurer, A. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of b-homophenylalanine based pyrrolidin-2-ylmethyl amides and sulfonamides as highly potent and selective inhibitors of dipeptidyl peptidase IV
To be Published
|
|
1J7M
 
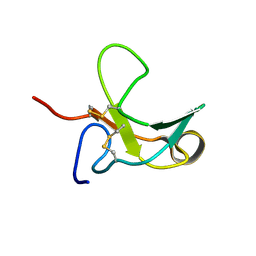 | | The Third Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Gehrmann, M, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-05-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct.
J.Biol.Chem., 276, 2001
|
|
3H6P
 
 | | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis | | Descriptor: | ESAT-6 LIKE PROTEIN ESXS, ESAT-6-like protein esxR, GLYCEROL | | Authors: | Chan, S, Arbing, M, Phan, T, Kaufmann, M, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis
To be Published
|
|
1SOZ
 
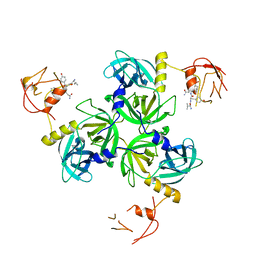 | | Crystal Structure of DegS protease in complex with an activating peptide | | Descriptor: | Protease degS, activating peptide | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1THF
 
 | |
3Q8J
 
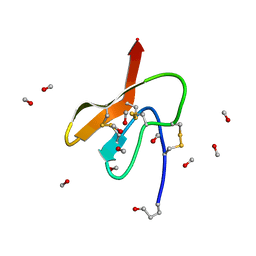 | | Crystal Structure of Asteropsin A from Marine Sponge Asteropus sp. | | Descriptor: | Asteropsin A, METHANOL | | Authors: | Bowling, J.J, Fronczek, F.R, Hamann, M.T, Li, H, Jung, J.H. | | Deposit date: | 2011-01-06 | | Release date: | 2012-04-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | An Uncommon Crystal Structure of a Marine Knottin Peptide from Asteropus sp.
To be published
|
|
2C0L
 
 | |
3EO5
 
 | | Crystal structure of the resuscitation promoting factor RpfB | | Descriptor: | Resuscitation-promoting factor rpfB | | Authors: | Ruggiero, A, Tizzano, B, Pedone, E, Pedone, C, Wilmanns, M, Berisio, R. | | Deposit date: | 2008-09-26 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the Resuscitation-Promoting Factor (DeltaDUF)RpfB from M. tuberculosis
J.Mol.Biol., 385, 2009
|
|
4G1O
 
 | |
3QP3
 
 | |
5LYW
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX; WITH AN INHIBITOR 6-((R)-2-o-Tolyloxymethyl-pyrrolidin-1-yl)-9H-purine | | Descriptor: | 6-[(2~{R})-2-[(2-methylphenoxy)methyl]pyrrolidin-1-yl]-7~{H}-purine, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Musil, D, Heinrich, T, Knoechel, T, Lehmann, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel reversible methionine aminopeptidase-2 (MetAP-2) inhibitors based on purine and related bicyclic templates.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5LDH
 
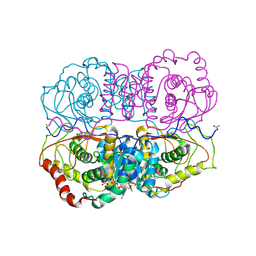 | |
1JCM
 
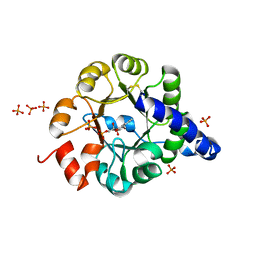 | | TRPC STABILITY MUTANT CONTAINING AN ENGINEERED DISULPHIDE BRIDGE AND IN COMPLEX WITH A CDRP-RELATED SUBSTRATE | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, INDOLE-3-GLYCEROL-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Ivens, A, Mayans, O, Szadkowski, H, Wilmanns, M, Kirschner, K. | | Deposit date: | 2001-06-10 | | Release date: | 2002-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stabilization of a (betaalpha)8-barrel protein by an engineered disulfide bridge.
Eur.J.Biochem., 269, 2002
|
|
3CT0
 
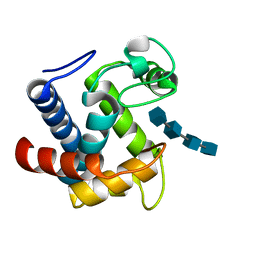 | |
7PSO
 
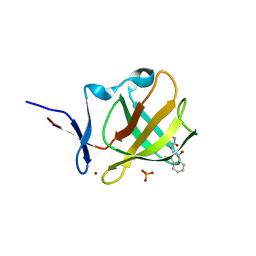 | |
4HUH
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15 (C-terminal truncation mutant 1-261). | | Descriptor: | Tail connector protein Gp15 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3CSR
 
 | |
3CT1
 
 | |
2BK8
 
 | |
3ZR4
 
 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
