6R1K
 
 | |
6R1W
 
 | |
6R1C
 
 | |
6R60
 
 | |
2XQH
 
 | | Crystal structure of an immunoglobulin-binding fragment of the trimeric autotransporter adhesin EibD | | 分子名称: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | 著者 | Leo, J.C, Lyskowski, A, Hartmann, M, Schwarz, H, Linke, D, Lupas, A.N, Goldman, A. | | 登録日 | 2010-09-02 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
2V8C
 
 | | Mouse Profilin IIa in complex with the proline-rich domain of VASP | | 分子名称: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | 著者 | Kursula, P, Downer, J, Witke, W, Wilmanns, M. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
3STJ
 
 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | 分子名称: | Protease degQ, peptide (UNK) | | 著者 | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | 登録日 | 2011-07-11 | | 公開日 | 2011-07-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
4HAR
 
 | |
4HUD
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15. | | 分子名称: | Tail connector protein Gp15 | | 著者 | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | 登録日 | 2012-11-02 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7001 Å) | | 主引用文献 | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3GVM
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | 分子名称: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | 著者 | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | 登録日 | 2013-08-30 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
1SOZ
 
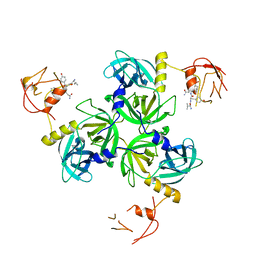 | | Crystal Structure of DegS protease in complex with an activating peptide | | 分子名称: | Protease degS, activating peptide | | 著者 | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | 登録日 | 2004-03-16 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VCW
 
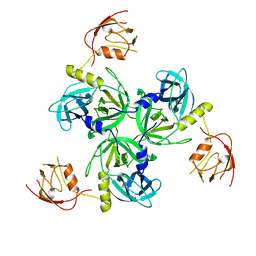 | | Crystal structure of DegS after backsoaking the activating peptide | | 分子名称: | Protease degS | | 著者 | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | 登録日 | 2004-03-16 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease.
Cell(Cambridge,Mass.), 117, 2004
|
|
1CWW
 
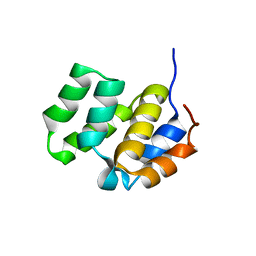 | | SOLUTION STRUCTURE OF THE CASPASE RECRUITMENT DOMAIN (CARD) FROM APAF-1 | | 分子名称: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | 著者 | Day, C.L, Dupont, C, Lackmann, M, Vaux, D.L, Hinds, M.G. | | 登録日 | 1999-08-26 | | 公開日 | 2000-01-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1.
Cell Death Differ., 6, 1999
|
|
2SNV
 
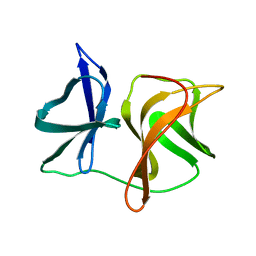 | |
5AF3
 
 | |
7PS9
 
 | |
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LN0
 
 | | Low dose Thaumatin - 760-800 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-08-02 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH7
 
 | | High dose Thaumatin - 760-800 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
7NAZ
 
 | |
5LH3
 
 | | High dose Thaumatin - 0-40 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
2BUB
 
 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (CD26) in Complex with a Reversed Amide Inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIPEPTIDYL PEPTIDASE 4, N-({(2S)-1-[(3R)-3-AMINO-4-(2-FLUOROPHENYL)BUTANOYL]PYRROLIDIN-2-YL}METHYL)BENZAMIDE | | 著者 | Nordhoff, S, Cerezo-Galvez, S, Feurer, A, Hill, O, Matassa, V.G, Metz, G, Rummey, C, Thiemann, M, Edwards, P.J. | | 登録日 | 2005-06-09 | | 公開日 | 2006-01-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | The reversed binding of beta-phenethylamine inhibitors of DPP-IV: X-ray structures and properties of novel fragment and elaborated inhibitors.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
4HUH
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15 (C-terminal truncation mutant 1-261). | | 分子名称: | Tail connector protein Gp15 | | 著者 | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | 登録日 | 2012-11-02 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
5J7O
 
 | |
