6CS4
 
 | |
6CS6
 
 | |
8DVX
 
 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
8DZL
 
 | | Structure of the K39Q mutant of rat somatic Cytochrome c at 1.36A | | Descriptor: | Cytochrome c, somatic, HEME C | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
4C10
 
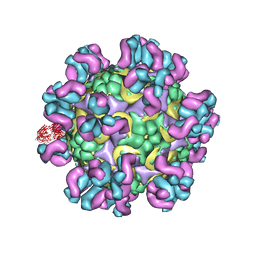 | | Cryo-EM reconstruction of empty enterovirus 71 in complex with a neutralizing antibody E19 | | Descriptor: | CHLORIDE ION, EV19 5 C1-6 F1 C11, SODIUM ION, ... | | Authors: | Plevka, P, Perera, R, Cardosa, J, Suksatu, A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2013-08-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CRP
 
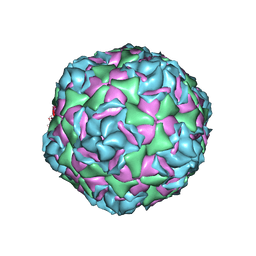 | |
6CSH
 
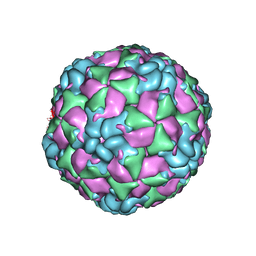 | |
6CRR
 
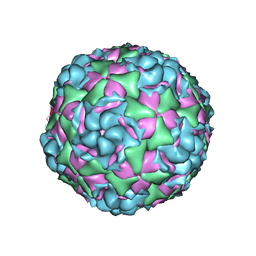 | |
6CSA
 
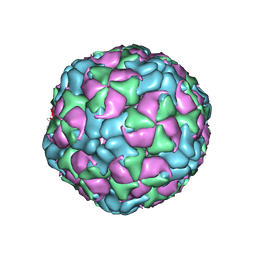 | |
6CSG
 
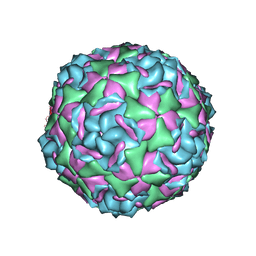 | |
6CS3
 
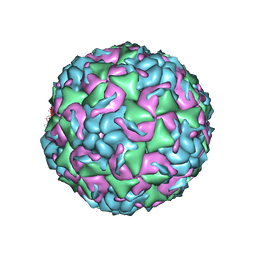 | |
6CS5
 
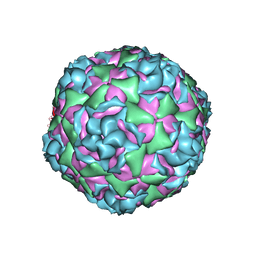 | |
4F14
 
 | |
4AZ0
 
 | | crystal structure of cathepsin a, complexed with 8a. | | Descriptor: | (S)-3-{[1-(2-Fluoro-phenyl)-5-hydroxy-1H-pyrazole-3-carbonyl]-amino}-3-o-tolyl-propionic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ruf, S, Buning, C, Schreuder, H, Horstick, G, Linz, W, Olpp, T, Pernerstorfer, J, Hiss, K, Kroll, K, Kannt, A, Kohlmann, M, Linz, D, Huebschle, T, Ruetten, H, Wirth, K, Schmidt, T, Sadowski, T. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Beta-Amino Acid Derivatives as Inhibitors of Cathepsin A.
J.Med.Chem., 55, 2012
|
|
4AUW
 
 | | CRYSTAL STRUCTURE OF THE BZIP HOMODIMERIC MAFB IN COMPLEX WITH THE C- MARE BINDING SITE | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C-MARE BINDING SITE (5'-D(*AP*TP*AP*AP*TP*GP*CP*TP* GP*AP*CP*GP*TP*CP*AP*GP*CP*AP*AP*TP*T)-3'), MERCURY (II) ION, ... | | Authors: | Textor, L.C, Holton, S, Wilmanns, M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of the mouse transcription factor MafB in complex with its DNA-recognition motif Cmare
Acta Crystallogr Sect F Struct Biol Cryst Commun., 63, 2007
|
|
3ZS4
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND PRFAR | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, PHOSPHORIC ACID MONO-[5-({[5-CARBAMOYL-3-(5-PHOSPHONOOXY-5-DEOXY-RIBOFURANOSYL)- 3H-IMIDAZOL-4-YLAMINO]-METHYL}-AMINO)-2,3,4-TRIHYDROXY-PENTYL] ESTER | | Authors: | Due, A.V, Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Phosphoribosyl Isomerase with Bound Prfar
To be Published
|
|
4CE5
 
 | | First crystal structure of an (R)-selective omega-transaminase from Aspergillus terreus | | Descriptor: | AT-OMEGATA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lyskowski, A, Gruber, C, Steinkellner, G, Schurmann, M, Schwab, H, Gruber, K, Steiner, K. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Plos One, 9, 2014
|
|
5OVU
 
 | | Cupriavidus metallidurans BPH | | Descriptor: | BETA-PROTEOBACTERIA PROTEASOME HOMOLOGUE, MALONATE ION | | Authors: | Fuchs, A.C.D, Albrecht, R, Martin, J, Hartmann, M.D. | | Deposit date: | 2017-08-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the bacterial proteasome homolog BPH reveals a tetradecameric double-ring complex with unique inner cavity properties.
J. Biol. Chem., 293, 2018
|
|
4EO2
 
 | |
8RDS
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDQ
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDT
 
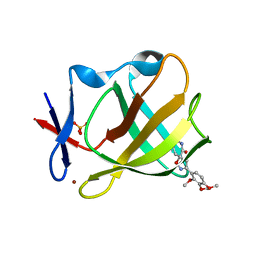 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
5OH4
 
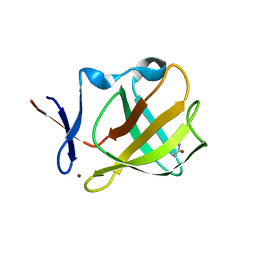 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH2
 
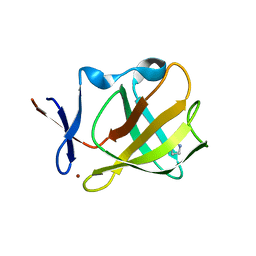 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OHB
 
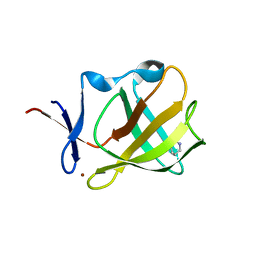 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidin-2-one (Valerolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
