7UY4
 
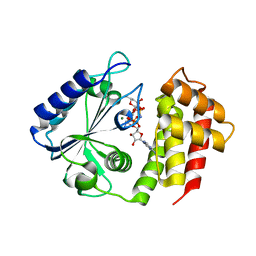 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
6B48
 
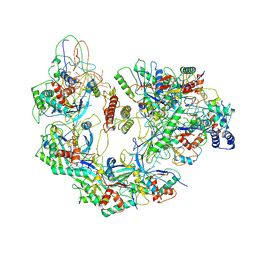 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6PPR
 
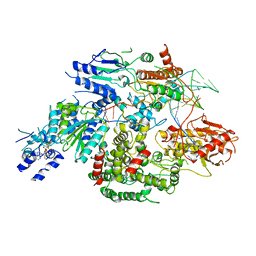 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5TR7
 
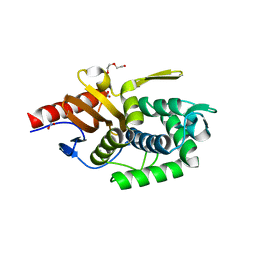 | | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961
To Be Published
|
|
6PPJ
 
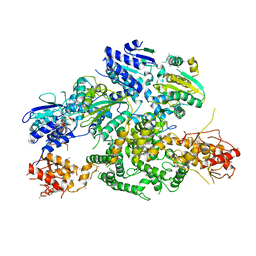 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2AJB
 
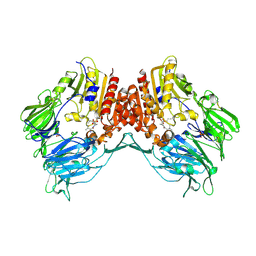 | | Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-methyl-L-valyl-L-prolyl-L-isoleucine, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
3C8T
 
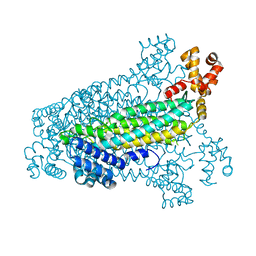 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
6V3V
 
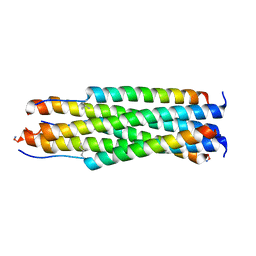 | |
4A0O
 
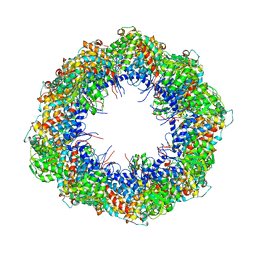 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
8J4W
 
 | |
6C2R
 
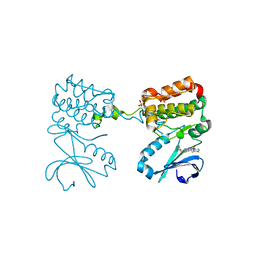 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
8U8L
 
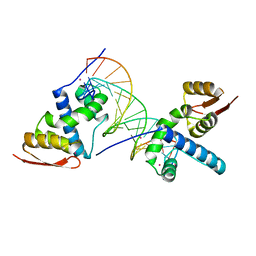 | | X-ray crystal structure of TEBP-2 MCD3 with ds DNA | | Descriptor: | CESIUM ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*CP*TP*TP*AP*GP*GP*CP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PPU
 
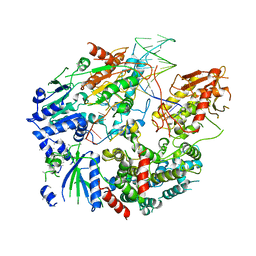 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5HFZ
 
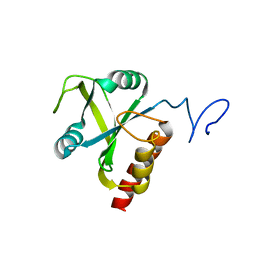 | | Mmi1 YTH domain | | Descriptor: | YTH domain-containing protein mmi1 | | Authors: | Chatterjee, D, Goldgur, Y, Shuman, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast.
Rna, 22, 2016
|
|
5UJW
 
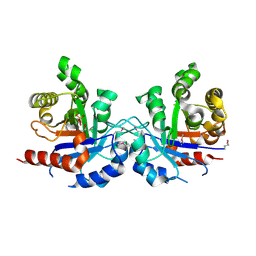 | | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CITRIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
6VAS
 
 | |
4A13
 
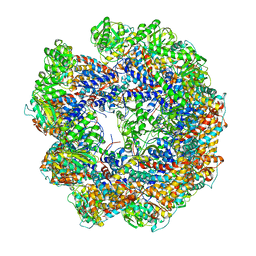 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
5NJW
 
 | | Crystal Structure of BJP-1 metallo beta-lactamase in complex with boric acid | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Blr6230 protein, ... | | Authors: | Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boric acid and acetate anion binding to subclass B3 metallo-Beta-lactamase BJP-1 provides clues for mechanism of action and inhibitor design
Inorg.Chim.Acta., 2017
|
|
5V9X
 
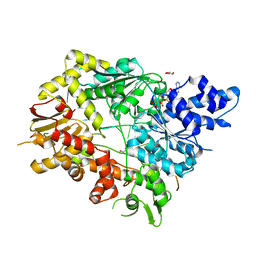 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | Descriptor: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | Deposit date: | 2017-03-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
5NGG
 
 | | Crystal structure of the subclass B3 metallo-beta-lactamase BJP-1 in complex with acetate anion | | Descriptor: | ACETATE ION, Blr6230 protein, ZINC ION | | Authors: | Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2017-03-17 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Boric acid and acetate anion binding to subclass B3 metallo-Beta-lactamase BJP-1 provides clues for mechanism of action and inhibitor design
Inorg.Chim.Acta., 2017
|
|
4A0W
 
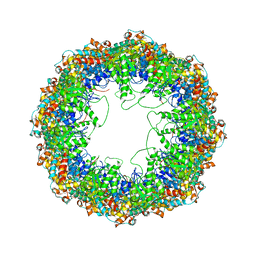 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
7T9L
 
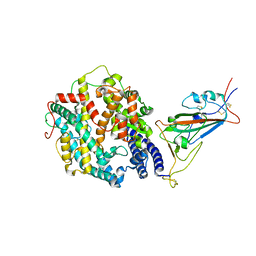 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
6CQA
 
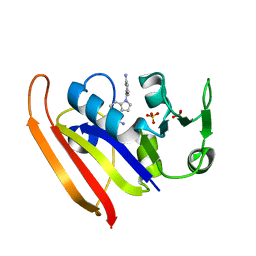 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
7T9J
 
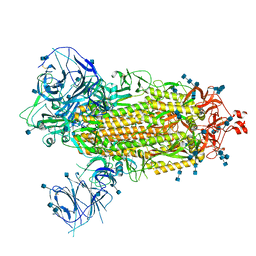 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
