8B4F
 
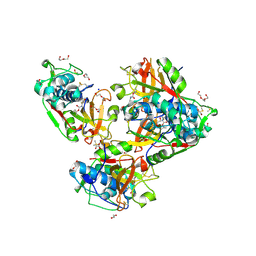 | | Crystal structure of human cathepsin L forming a thiohemiacetal with N-Boc-2-aminoacetaldehyde | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
1IN3
 
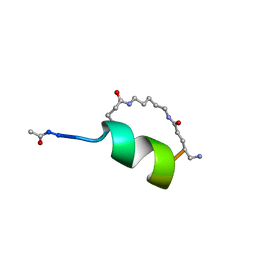 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
7YMQ
 
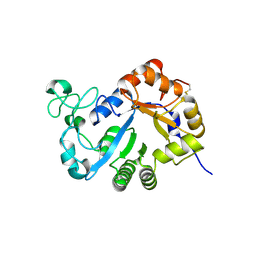 | | Crystal structure of lysoplasmalogen specific phopholipase D, F211L mutant | | Descriptor: | Lysoplasmalogenase | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
7YMR
 
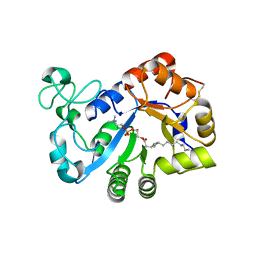 | | Complex structure of lysoplasmalogen specific phopholipase D, F211L mutant with LPC | | Descriptor: | Lysoplasmalogenase, [(2~{R})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{5}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
7YMP
 
 | | Crystal structure of lysoplasmalogen specific phospholipase D | | Descriptor: | Lysoplasmalogenase | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
1MRS
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH 5-CH2OH DEOXYURIDINE MONOPHOSPHATE | | Descriptor: | 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
3PZ2
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 and lipid substrate GGPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3Q5D
 
 | |
2BN7
 
 | | Mn substituted E. coli Aminopeptidase P in complex with product and Zn | | Descriptor: | CITRATE ANION, LEUCINE, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2BHB
 
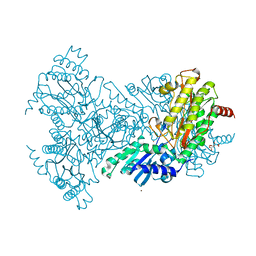 | | Zn substituted E. coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-08 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
8C77
 
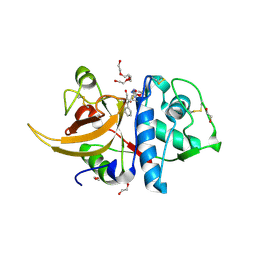 | | Human cathepsin L after reaction with the thiocarbazate inhibitor CID 16725315 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6VLN
 
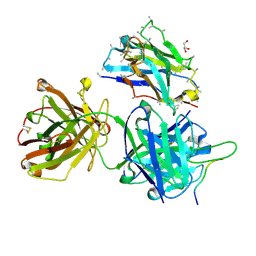 | | Crystal structure of 4498 Fab in complex with circumsporozoite protein DND3 and anti-Kappa VHH domain | | Descriptor: | 4498 Fab heavy chain, 4498 Fab light chain, Circumsporozoite protein, ... | | Authors: | Thai, E, Scally, S.W, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
7ZNF
 
 | |
8CJF
 
 | | AetF, a single-component flavin-dependent tryptophan halogenase, in complex with 5-bromo-L-tryptophan | | Descriptor: | 5-bromo-L-tryptophan, AetF, CHLORIDE ION, ... | | Authors: | Gafe, S, Niemann, H.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of regioselective tryptophan dibromination by the single-component flavin-dependent halogenase AetF.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CJD
 
 | | AetF, a single-component flavin-dependent tryptophan halogenase | | Descriptor: | 1,2-ETHANEDIOL, AetF, CALCIUM ION, ... | | Authors: | Gafe, S, Niemann, H.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of regioselective tryptophan dibromination by the single-component flavin-dependent halogenase AetF.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CJG
 
 | | AetF, a single-component flavin-dependent tryptophan halogenase, in complex with 7-bromo-L-tryptophan | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-L-tryptophan, AetF, ... | | Authors: | Gafe, S, Niemann, H.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of regioselective tryptophan dibromination by the single-component flavin-dependent halogenase AetF.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CJE
 
 | |
1P4S
 
 | |
7PSC
 
 | | Crystal structure of the disease-causing I358T mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nemes-Nikodem, E, Szabo, E, Zambo, Z, Vass, K.R, Taberman, H, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2021-09-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural and Biochemical Investigation of Selected Pathogenic Mutants of the Human Dihydrolipoamide Dehydrogenase.
Int J Mol Sci, 24, 2023
|
|
3PCY
 
 | | THE CRYSTAL STRUCTURE OF MERCURY-SUBSTITUTED POPLAR PLASTOCYANIN AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | MERCURY (II) ION, PLASTOCYANIN | | Authors: | Church, W.B, Guss, J.M, Potter, J.J, Freeman, H.C. | | Deposit date: | 1985-12-10 | | Release date: | 1986-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of mercury-substituted poplar plastocyanin at 1.9-A resolution.
J.Biol.Chem., 261, 1986
|
|
6GF3
 
 | | Tubulin-Jerantinine B acetate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Smedley, C.J, Stanley, P.A, Qazzaz, M.E, Prota, A.E, Olieric, N, Collins, H, Eastman, H, Barrow, A.S, Lim, K.-H, Kam, T.-S, Smith, B.J, Duivenvoorden, H.M, Parker, B.S, Bradshaw, T.D, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Syntheses of (-)-Jerantinines A & E and Structural Characterisation of the Jerantinine-Tubulin Complex at the Colchicine Binding Site.
Sci Rep, 8, 2018
|
|
3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2CBP
 
 | | CUCUMBER BASIC PROTEIN, A BLUE COPPER PROTEIN | | Descriptor: | COPPER (II) ION, CUCUMBER BASIC PROTEIN | | Authors: | Guss, J.M, Freeman, H.C. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a phytocyanin, the basic blue protein from cucumber, refined at 1.8 A resolution.
J.Mol.Biol., 262, 1996
|
|
6HZJ
 
 | | Apo structure of TP domain from clinical penicillin-resistant mutant Neisseria gonorrhoea strain 6140 Penicillin-Binding Protein 2 (PBP2) | | Descriptor: | Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
