4NH5
 
 | |
1NCJ
 
 | | N-CADHERIN, TWO-DOMAIN FRAGMENT | | Descriptor: | CALCIUM ION, PROTEIN (N-CADHERIN), URANYL (VI) ION | | Authors: | Tamura, K, Shan, W.-S, Hendrickson, W.A, Colman, D.R, Shapiro, L. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function analysis of cell adhesion by neural (N-) cadherin.
Neuron, 20, 1998
|
|
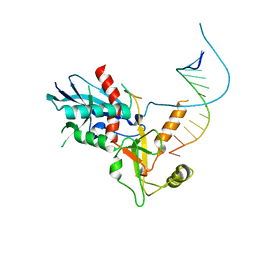
1QZ7
 
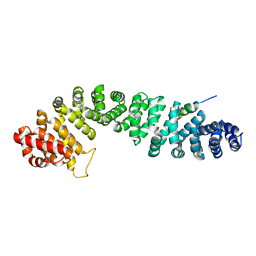 | | Beta-catenin binding domain of Axin in complex with beta-catenin | | Descriptor: | Axin, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a beta-catenin/Axin complex suggests a mechanism for the {beta}-catenin destruction complex
GENES DEV., 17, 2003
|
|
3FWO
 
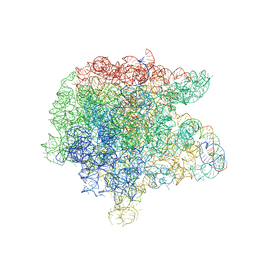 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
6B11
 
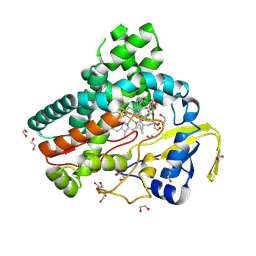 | | TylHI in complex with native substrate 23-deoxy-5-O-mycaminosyl-tylonolide (23-DMTL) | | Descriptor: | (4R,5S,6S,7R,9R,11E,13E,15S,16R)-16-ethyl-4-hydroxy-5,9,13,15-tetramethyl-2,10-dioxo-7-(2-oxoethyl)-1-oxacyclohexadeca-11,13-dien-6-yl 3,6-dideoxy-3-(dimethylamino)-beta-D-glucopyranoside, 1,2-ETHANEDIOL, 20-oxo-5-O-mycaminosyltylactone 23-monooxygenase, ... | | Authors: | DeMars, M.D, Sherman, D.H, Podust, L.M. | | Deposit date: | 2017-09-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | TylHI in complex with native substrate 23-deoxy-5-O-mycaminosyl-tylonolide (23-DMTL)
To Be Published
|
|
4OYK
 
 | |
6V65
 
 | | Crystal structure of KRAS(GMPPNP)-NF1(GRD)-SPRED1 complex | | Descriptor: | FORMIC ACID, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Yan, W, Simanshu, D.K. | | Deposit date: | 2019-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR.
Cell Rep, 32, 2020
|
|
1DPS
 
 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1KFW
 
 | |
1HFH
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1KIB
 
 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJN
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
3GYT
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 4 | | Descriptor: | (14beta,17alpha,25R)-3-oxocholest-4-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1NHN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NHM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NEU
 
 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
6W9O
 
 | |
6W9R
 
 | |
1O0S
 
 | | Crystal Structure of Ascaris suum Malic Enzyme Complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD-dependent malic enzyme, TARTRONATE | | Authors: | Rao, G.S, Coleman, D.E, Karsten, W.E, Cook, P.F, Harris, B.G. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
J.Biol.Chem., 278, 2003
|
|
