3G12
 
 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3BGE
 
 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | Descriptor: | Predicted ATPase, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
3B5M
 
 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
6VGI
 
 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
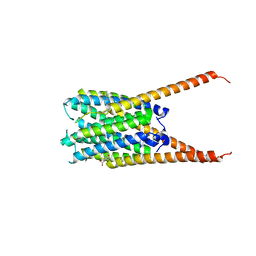 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VL4
 
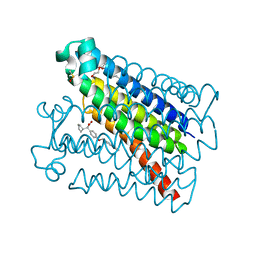 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
2QH5
 
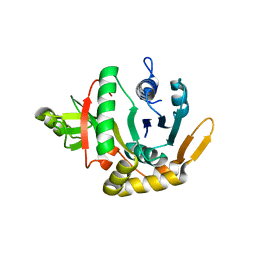 | | Crystal structure of mannose-6-phosphate isomerase from Helicobacter pylori | | Descriptor: | Mannose-6-phosphate isomerase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Iizuka, M, Maletic, M, Wasserman, S.R, Koss, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mannose-6-phosphate isomerase from Helicobacter pylori.
To be Published
|
|
2R1F
 
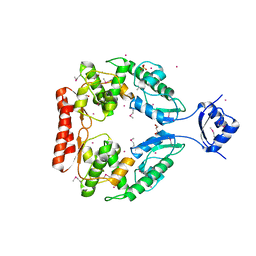 | | Crystal structure of predicted aminodeoxychorismate lyase from Escherichia coli | | Descriptor: | CADMIUM ION, GLYCEROL, Predicted aminodeoxychorismate lyase, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Rutter, M, Lau, C, Maletic, M, Smith, D, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Predicted Aminodeoxychorismate Lyase from Escherichia coli.
To be Published
|
|
2RDX
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Ozyurt, S, Gilmore, M, Lau, C, Maletic, M, Gheyi, T, Wasserman, S.R, Koss, J, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM.
To be Published
|
|
6MNH
 
 | | ULK1 Unc-51 like autophagy activating kinase in complex with inhibitor BTC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-[(2R)-3-methylbutan-2-yl]-1H-benzotriazole-6-carboxamide, ... | | Authors: | Hendle, J, Sauder, J.M, Hickey, M.J, Rauch, C.T, Maletic, M, Schwinn, K.D. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Idea2Data: Toward a New Paradigm for Drug Discovery.
Acs Med.Chem.Lett., 10, 2019
|
|
3NEK
 
 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
2OOG
 
 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | Authors: | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I1Y
 
 | | Crystal structure of the phosphatase domain of human PTP IA-2 | | Descriptor: | GLYCEROL, Receptor-type tyrosine-protein phosphatase | | Authors: | Faber-Barata, J, Patskovsky, Y, Alvarado, J, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Maletic, M, Rooney, I, Bain, K.T, Freeman, M, Russell, J.C, Thompson, D.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3BY5
 
 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | Descriptor: | Cobalamin biosynthesis protein, SULFATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3BVC
 
 | | Crystal structure of uncharacterized protein Ism_01780 from Roseovarius nubinhibens ISM | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Uncharacterized protein Ism_01780 | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Rutter, M, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-06 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an uncharacterized protein Ism_01780 from Roseovarius nubinhibens.
To be Published
|
|
3CE2
 
 | | Crystal structure of putative peptidase from Chlamydophila abortus | | Descriptor: | Putative peptidase, ZINC ION | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Eberle, M, Maletic, M, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative peptidase from Chlamydophila abortus.
To be Published
|
|
3E05
 
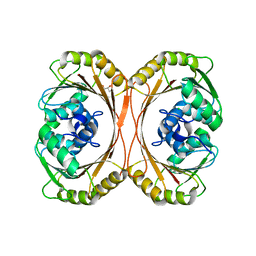 | | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase FROM Geobacter metallireducens GS-15 | | Descriptor: | CHLORIDE ION, GLYCEROL, Precorrin-6Y C5,15-methyltransferase (Decarboxylating) | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Dickey, M, Hu, S, Maletic, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF Precorrin-6y C5,15-methyltransferase from Geobacter metallireducens
To be Published
|
|
3DJC
 
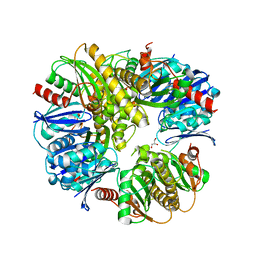 | | CRYSTAL STRUCTURE OF PANTOTHENATE KINASE FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | GLYCEROL, Type III pantothenate kinase | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Dickey, M, Logan, C, Wasserman, S, Maletic, M, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantothenate Kinase from Legionella Pneumophila
To be Published
|
|
3EGO
 
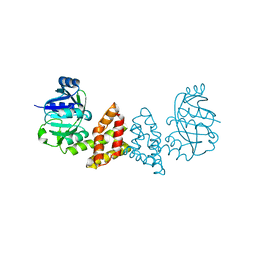 | | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis | | Descriptor: | Probable 2-dehydropantoate 2-reductase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-11 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Probable 2-dehydropantoate 2-reductase panE from Bacillus Subtilis
To be published
|
|
3DO9
 
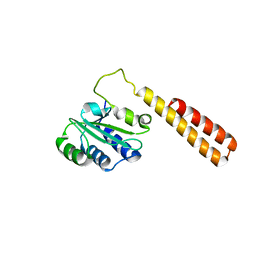 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3CMG
 
 | | Crystal structure of putative beta-galactosidase from Bacteroides fragilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Ramagopal, U.A, Rutter, M, Toro, R, Hu, S, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative beta-galactosidase from Bacteroides fragilis.
To be published
|
|
3EWB
 
 | | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes | | Descriptor: | 2-isopropylmalate synthase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes.
To be Published
|
|
