7YZE
 
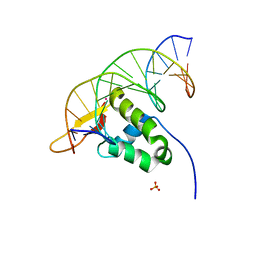 | |
7YZF
 
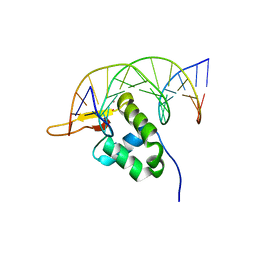 | |
6FQP
 
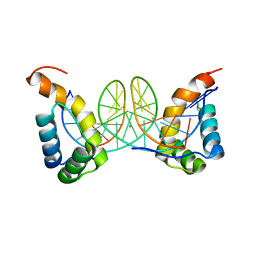 | |
6FQQ
 
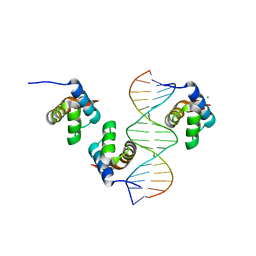 | |
7Z9T
 
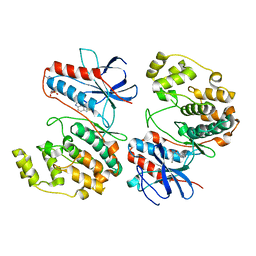 | | Crystal structure of p38alpha C162S in complex with ATPgS and CAS 2094667-81-7 (in catalytic site, Y35 out), P 1 21 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)benzenesulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-03-21 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
6H3R
 
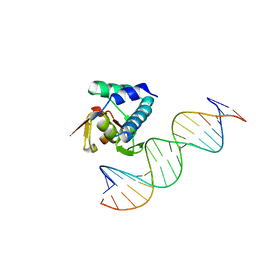 | |
6FZS
 
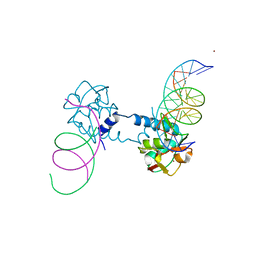 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6FZT
 
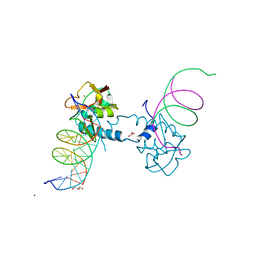 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
2JUP
 
 | | FBP28WW2 domain in complex with the PPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-01 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2JUC
 
 | | URN1 FF domain yeast | | Descriptor: | Pre-mRNA-splicing factor URN1 | | Authors: | Bonet, R, Ramirez-Espain, X, Macias, M.J. | | Deposit date: | 2007-08-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the yeast URN1 splicing factor FF domain: Comparative analysis of charge distributions in FF domain structures-FFs and SURPs, two domains with a similar fold.
Proteins, 73, 2008
|
|
5MEZ
 
 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MEY
 
 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
7Z6I
 
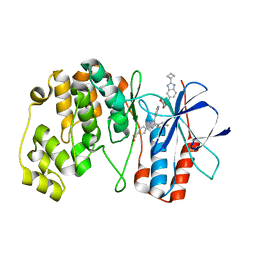 | | Crystal structure of p38alpha C162S in complex with SB20358 and CAS 2094667-81-7 (behind catalytic site; Y35 in), P 21 21 21 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-[4-[methyl(oxidanyl)-$l^{3}-sulfanyl]phenyl]-1~{H}-imidazol-5-yl]pyridine, Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)benzenesulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
5MF0
 
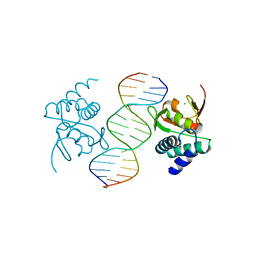 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5OD6
 
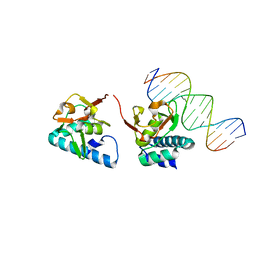 | |
5ODG
 
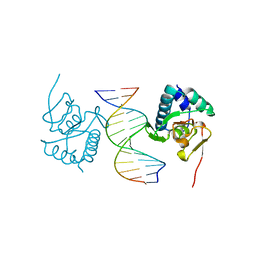 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
2MPT
 
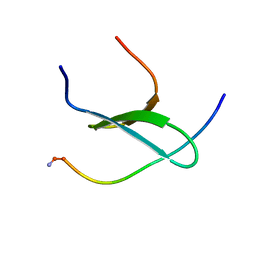 | | WW3 domain of Nedd4L in complex with its HECT domain PY motif | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Escobedo, A, Macias, M.J, Gomes, T, Aragon, E, Martin-Malpartida, P, Ruiz, L. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Activation and Degradation Mechanisms of the E3 Ubiquitin Ligase Nedd4L.
Structure, 22, 2014
|
|
2O4G
 
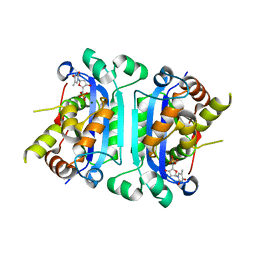 | | Structure of TREX1 in complex with a nucleotide | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1 | | Authors: | Brucet, M, Macias, M.J, Fita, I, Celada, A. | | Deposit date: | 2006-12-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains.
J.Biol.Chem., 282, 2007
|
|
2O4I
 
 | | Structure of TREX1 in complex with DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*GP*GP*CP*AP*GP*GP*AP*AP*CP*CP*CP*CP*TP*CP*CP*TP*CP*CP*CP*CP*T)-3', Three prime repair exonuclease 1 | | Authors: | Brucet, M, Macias, M.J, Fita, I, Celada, A. | | Deposit date: | 2006-12-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains.
J.Biol.Chem., 282, 2007
|
|
