1X7Y
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPE
 
 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPA
 
 | | Transcription factor FosB/JunD bZIP domain | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPF
 
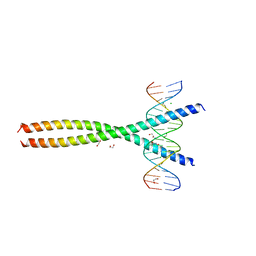 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
3V1B
 
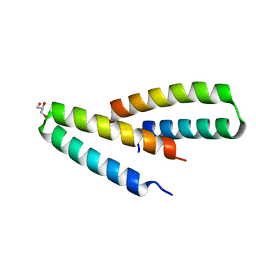 | | Crystal structure of de novo designed MID1-apo2 | | Descriptor: | Computational design, MID1-apo2, GLYCEROL | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
1U5Q
 
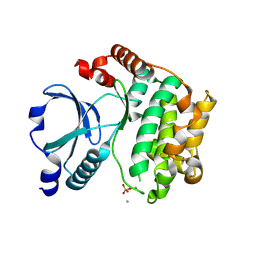 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | CALCIUM ION, serine/threonine protein kinase TAO2 | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
STRUCTURE, 12, 2004
|
|
1U5R
 
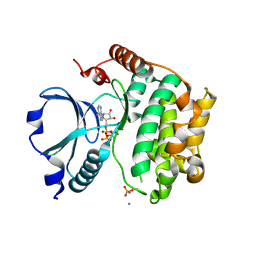 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
Structure, 12, 2004
|
|
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
3U3B
 
 | |
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1C
 
 | | Crystal structure of de novo designed MID1-zinc | | Descriptor: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
 | | Crystal structure of de novo designed MID1-cobalt | | Descriptor: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
2LCH
 
 | | Solution NMR Structure of a Protein With a Redesigned Hydrophobic Core, Northeast Structural Genomics Consortium Target OR38 | | Descriptor: | Protein OR38 | | Authors: | Mills, J.L, Murphy, G, Miley, M, Machius, M, Kuhlman, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Protein With a Redesigned Hydrophobic Core, Northeast Structural Genomics Consortium Target OR38
To be Published
|
|
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1U3D
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2OIX
 
 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
1U3C
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
2LSE
 
 | | Solution NMR Structure of De Novo Designed Four Helix Bundle Protein, Northeast Structural Genomics Consortium (NESG) Target OR188 | | Descriptor: | Four Helix Bundle Protein | | Authors: | Sathyamoorthy, B, Pulavarti, S, Murphy, G, Mills, J.L, Eletski, A, Der, B.S, Machius, M.C, Kuhlman, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Denovo Design Four Helix Bundle Protein, Northeast Structural Genomics Consortium Target OR188
To be Published
|
|
