6Q90
 
 | | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea
To Be Published
|
|
5UZ8
 
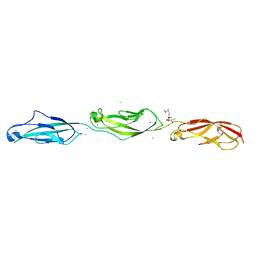 | | Crystal Structure of Mouse Cadherin-23 EC22-24 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Patel, A, Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
6Q4R
 
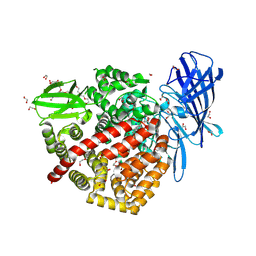 | | High-resolution crystal structure of ERAP1 with bound phosphinic transition-state analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Neu, M, Rowland, P, Stratikos, E. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of Endoplasmic Reticulum Aminopeptidase 1 with Bound Phosphinic Transition-State Analogue Inhibitor.
Acs Med.Chem.Lett., 10, 2019
|
|
1S0V
 
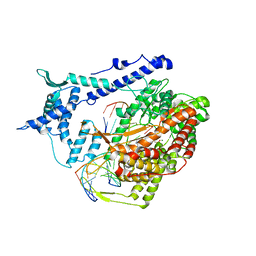 | | Structural basis for substrate selection by T7 RNA polymerase | | Descriptor: | 5'-D(*G*GP*GP*AP*AP*TP*CP*GP*AP*TP*AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*C)-3', 5'-R(*AP*AP*CP*U*GP*CP*GP*GP*CP*GP*AP*U)-3', ... | | Authors: | Temiakov, D, Patlan, V, Anikin, M, McAllister, W.T, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selection by t7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
6QCI
 
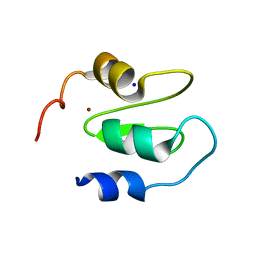 | | Structure of XIAP-BIR1 V86E mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Sorrentino, L, Cossu, F, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
5V11
 
 | |
2XDH
 
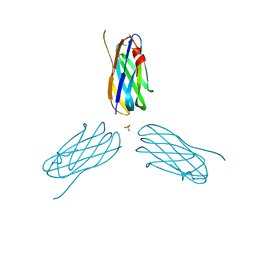 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
6Q6C
 
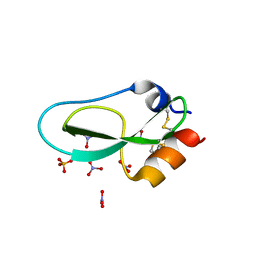 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | 1,2-ETHANEDIOL, Kunitz-type conkunitzin-S1, NITRATE ION, ... | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7MYT
 
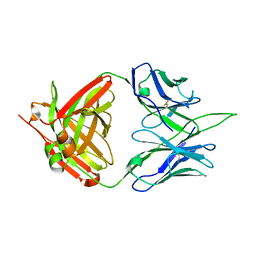 | |
6QD1
 
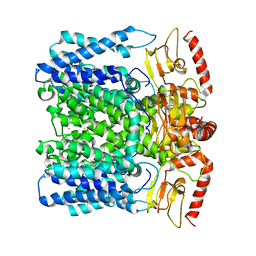 | | MloK1 model from single particle analysis of 2D crystals, class 5 (intermediate compact conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
5V2X
 
 | | Ethylene forming enzyme in complex with manganese and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
7JIS
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
7JJD
 
 | |
3TEN
 
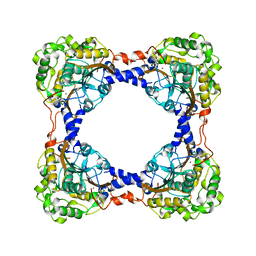 | | Holo form of carbon disulfide hydrolase | | Descriptor: | CS2 hydrolase, ZINC ION | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
5V8F
 
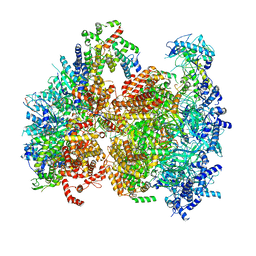 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QK0
 
 | |
7JGV
 
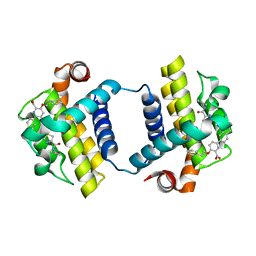 | | CRYSTAL STRUCTURE OF BCL-XL IN COMPLEX WITH COMPOUND 1620116, CRYSTAL FORM 2 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7MZX
 
 | |
2XS4
 
 | | Structure of karilysin catalytic MMP domain in complex with magnesium | | Descriptor: | CHLORIDE ION, KARILYSIN PROTEASE, MAGNESIUM ION, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
7JGW
 
 | | Crystal structure of BCL-XL in complex with COMPOUND 1620116, CRYSTAL FORM 1 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
5OS7
 
 | | The crystal structure of CK2alpha in complex with compound 4 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
2O11
 
 | |
6CD1
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), complexes with reaction intermediates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6QGA
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
