6L01
 
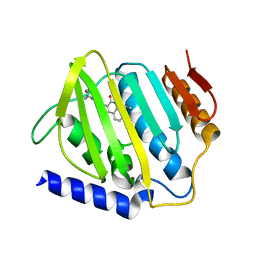 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 2-[3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]phenyl]ethanoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
9FEH
 
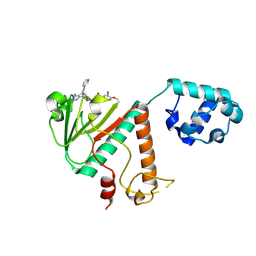 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | Descriptor: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | Authors: | Zilecka, E, Klima, M, Boura, E. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
279D
 
 | |
4WAF
 
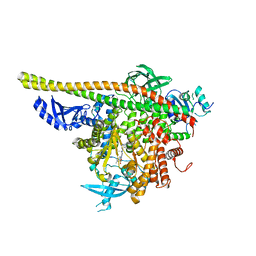 | | Crystal Structure of a novel tetrahydropyrazolo[1,5-a]pyrazine in an engineered PI3K alpha | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Drug Design of Novel Potent and Selective Tetrahydropyrazolo[1,5-a]pyrazines as ATR Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
1ZZE
 
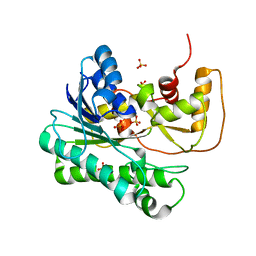 | | X-ray Structure of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor | | Descriptor: | Aldehyde reductase II, SULFATE ION | | Authors: | Kamitori, S, Iguchi, A, Ohtaki, A, Yamada, M, Kita, K. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
290D
 
 | |
213D
 
 | |
5OF2
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9FAB
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
246D
 
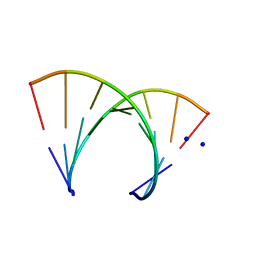 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2A3V
 
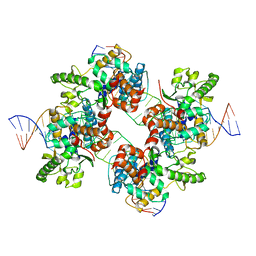 | | Structural basis for broad DNA-specificity in integron recombination | | Descriptor: | DNA (31-MER), DNA (34-MER), site-specific recombinase IntI4 | | Authors: | MacDonald, D, Demarre, G, Bouvier, M, Mazel, D, Gopaul, D.N. | | Deposit date: | 2005-06-27 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for broad DNA-specificity in integron recombination.
Nature, 440, 2006
|
|
2A50
 
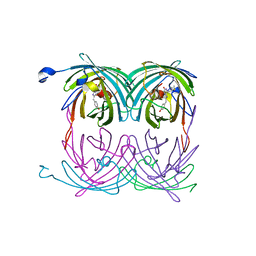 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5OLK
 
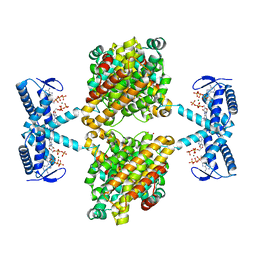 | | Crystal structure of the ATP-cone-containing NrdB from Leeuwenhoekiella blandensis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Hasan, M, Grinberg, A.R, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Elife, 7, 2018
|
|
2A08
 
 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
2A6O
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
2A0P
 
 | |
2A37
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (DRKN SH3 DOMAIN) | | Descriptor: | Protein E(sev)2B | | Authors: | Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
8Y9X
 
 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
8Y7E
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U12 snRNP part | | Descriptor: | PHD finger-like domain-containing protein 5A, Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
2A9K
 
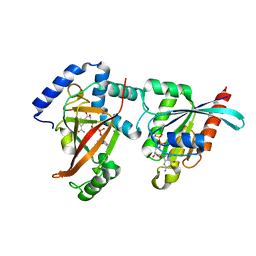 | | Crystal structure of the C3bot-NAD-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
2A4W
 
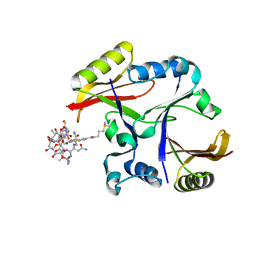 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
9F8G
 
 | |
8Y6H
 
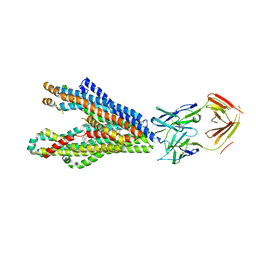 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
2A5L
 
 | | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Trp repressor binding protein WrbA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa
To be Published
|
|
