3OWC
 
 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
6KAH
 
 | | Crosslinked alpha(Ni)-beta(Fe-CO) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-23 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAQ
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAU
 
 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3OP3
 
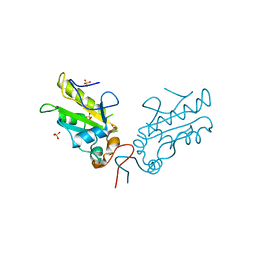 | | Crystal Structure of Cell Division Cycle 25C Protein Isoform A from Homo sapiens | | Descriptor: | M-phase inducer phosphatase 3, SULFATE ION | | Authors: | Kim, Y, Weger, A, Hatzos, C, Savitsky, P, Johansson, C, Ball, L, Barr, A, Vollmar, M, Muniz, J, Weigelt, J, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, von Delft, F, Knapp, S, Joachimiak, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structure of Cell Division Cycle 25C Protein Isoform A from Homo sapiens
TO BE PUBLISHED
|
|
3OZ3
 
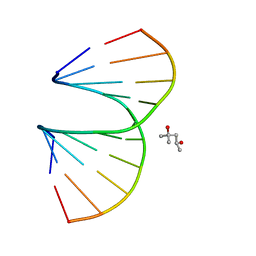 | | Vinyl Carbocyclic LNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*(UVX)P*AP*CP*GP*C)-3') | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
3OP9
 
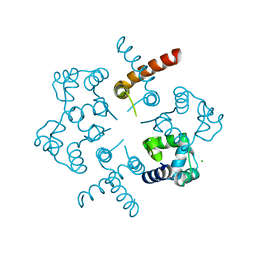 | |
6KDU
 
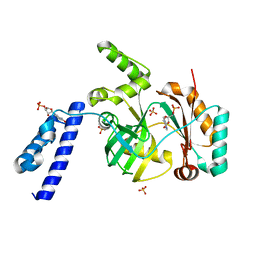 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3P0R
 
 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
3P36
 
 | | Polo-like kinase I Polo-box domain in complex with DPPLHSpTA phosphopeptide from PBIP1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Sledz, P, Stubbs, C.J, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P58
 
 | |
6O39
 
 | | Crystal structure of Frizzled 5 CRD in complex with F2.I Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Raman, S, Beilschmidt, M, Fransson, J, Julien, J.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided design fine-tunes pharmacokinetics, tolerability, and antitumor profile of multispecific frizzled antibodies.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3OSJ
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 254-400) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | NITRATE ION, Phycobilisome LCM core-membrane linker polypeptide, SODIUM ION | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-09 | | Release date: | 2010-10-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209C
To be published
|
|
3OL0
 
 | |
6K7J
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7Y
 
 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
3QPB
 
 | | Crystal Structure of Streptococcus Pyogenes Uridine Phosphorylase Reveals a Subclass of the NP-I Superfamily | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, URACIL, Uridine phosphorylase | | Authors: | Tran, T.H, Christoffersen, S, Parker, W.B, Piskur, J, Serra, I, Terreni, M, Ealick, S.E. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Streptococcus pyogenes Uridine Phosphorylase Reveals a Distinct Subfamily of Nucleoside Phosphorylases.
Biochemistry, 50, 2011
|
|
6K9K
 
 | | Monomeric human ATM (Ataxia telangiectasia mutated) kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Yuriy, C, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6KME
 
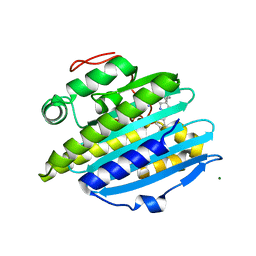 | | Crystal structure of phytochromobilin synthase from tomato in complex with biliverdin | | Descriptor: | BILIVERDINE IX ALPHA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of phytochromobilin synthase in complex with biliverdin IX alpha , a key enzyme in the biosynthesis of phytochrome.
J.Biol.Chem., 295, 2020
|
|
3QTD
 
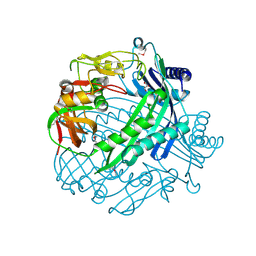 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
6KEV
 
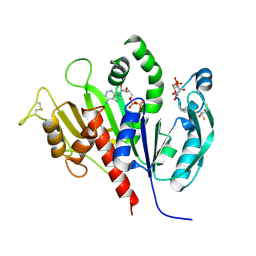 | | Reduced phosphoribulokinase from Synechococcus elongatus PCC 7942 complexed with adenosine diphosphate and glucose 6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.506139 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
3QV4
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Shukla, P.K, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution
To be Published
|
|
3QWM
 
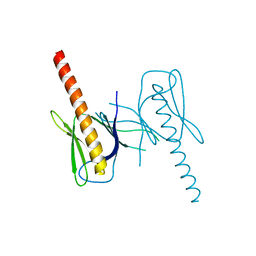 | | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a | | Descriptor: | IQ motif and SEC7 domain-containing protein 1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Feller, S, Janning, M, Sabe, H, Krojer, T, Chaikuad, A, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a
to be published
|
|
3QMN
 
 | | Crystal Structure of 4'-Phosphopantetheinyl Transferase AcpS from Vibrio cholerae O1 biovar eltor | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Kim, Y, Halavaty, A.S, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-04 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
