6K7H
 
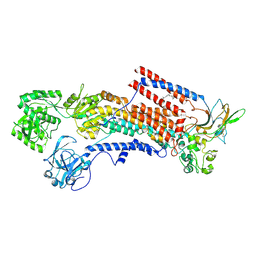 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
3QI3
 
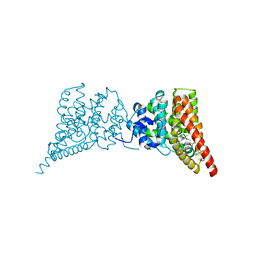 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
6K2P
 
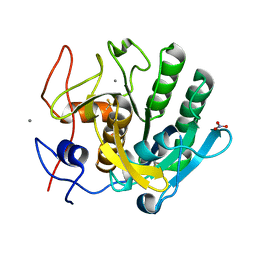 | |
6K2W
 
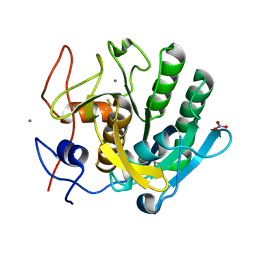 | |
3QKL
 
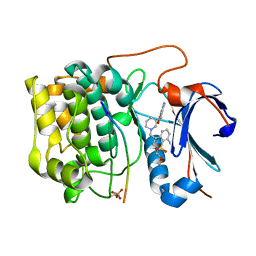 | | Spirochromane Akt Inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta, N-{(2S)-3-[(3S)-8',9'-dihydro-1H,3'H-spiro[piperidine-3,7'-pyrano[3,2-e]indazol]-1-yl]-2-hydroxypropyl}-N-(2-ethoxyethyl)-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QIS
 
 | | Recognition of the F&H motif by the Lowe Syndrome protein OCRL | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1, Protein FAM109A | | Authors: | Pirruccello, M, Swan, L.E, Folta-Stogniew, E, De Camilli, P. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of the F&H motif by the Lowe syndrome protein OCRL.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6K36
 
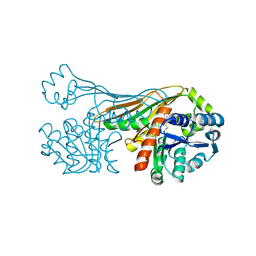 | |
3QJG
 
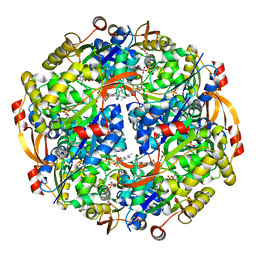 | | Epidermin biosynthesis protein EpiD from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Epidermin biosynthesis protein EpiD, FLAVIN MONONUCLEOTIDE | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Epidermin biosynthesis protein EpiD from Staphylococcus aureus.
To be Published
|
|
3QO3
 
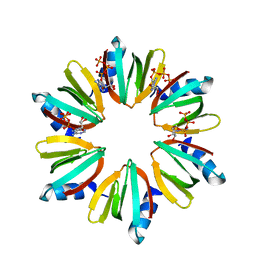 | | Crystal structure of Escherichia coli Hfq, in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Hammele, H, Kloiber, K, Sjoeblom, B, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq
Plos One, 7, 2012
|
|
6KA0
 
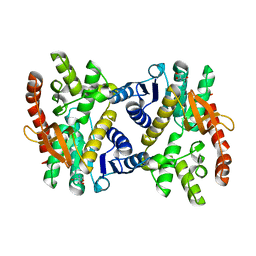 | | Silver-bound E.coli Malate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
6QBH
 
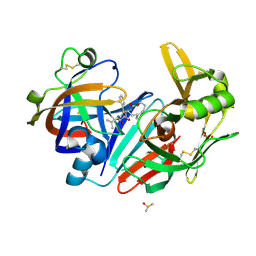 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 33 | | Descriptor: | (4~{S},5~{S},9~{S})-5-oxidanyl-4-(phenylmethyl)-9-propan-2-yl-1-oxa-3,8,11-triazacyclodocosane-2,7,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
3QVE
 
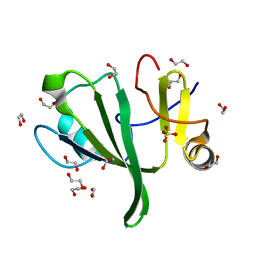 | | Crystal structure of human HMG box-containing protein 1, HBP1 | | Descriptor: | 1,2-ETHANEDIOL, HMG box-containing protein 1, SULFATE ION | | Authors: | Chaikuad, A, Krysztofinska, E, Cocking, R, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of human HMG box-containing protein 1, HBP1
To be Published
|
|
3Q5O
 
 | |
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6KAE
 
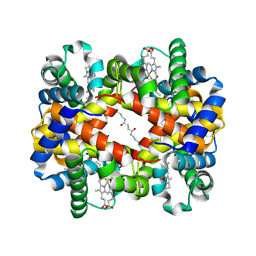 | | Crosslinked alpha(Fe-CO)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Light | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAO
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 95 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-23 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QD4
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl{(3R,5R)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-5-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, SULFATE ION, tert-butyl {(3R,5R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-5-methylpiperidin-3-yl}carbamate | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3Q8F
 
 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3Q8X
 
 | | Structure of a toxin-antitoxin system bound to its substrate | | Descriptor: | Antidote of epsilon-zeta postsegregational killing system, SULFATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, ... | | Authors: | Mutschler, H, Gebhardt, M, Shoeman, R.L, Meinhart, A. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis.
Plos Biol., 9, 2011
|
|
3QDC
 
 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the active state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-18 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
6KAR
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Light | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QBA
 
 | | Reintroducing Electrostatics into Macromolecular Crystallographic Refinement: Z-DNA (X-ray) | | Descriptor: | Z-DNA | | Authors: | Fenn, T.D, Schnieders, M.J, Mustyakimov, M, Wu, C, Langan, P, Pande, V.S, Brunger, A.T. | | Deposit date: | 2011-01-12 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.53 Å), X-RAY DIFFRACTION | | Cite: | Reintroducing electrostatics into macromolecular crystallographic refinement: application to neutron crystallography and DNA hydration.
Structure, 19, 2011
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QD3
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
