6EM8
 
 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
3ZKL
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with xylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, PUTATIVE SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Ejby, M, Vujicic-Zagar, A, Fredslund, F, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
1H72
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH HSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HOMOSERINE KINASE, L-HOMOSERINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
6TMY
 
 | | Crystal structure of isoform CBd of the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus | | Descriptor: | CHLORIDE ION, Phospholipase A2 crotoxin basic subunit CBc, SODIUM ION, ... | | Authors: | Nemecz, D, Ostrowski, M, Saul, F.A, Faure, G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Isoform CBd of the Basic Phospholipase A 2 Subunit of Crotoxin: Description of the Structural Framework of CB for Interaction with Protein Targets.
Molecules, 25, 2020
|
|
3ZJK
 
 | | crystal structure of Ttb-gly F401S mutant | | Descriptor: | BETA GLYCOSIDASE, CHLORIDE ION, GLYCEROL | | Authors: | Teze, D, Tran, V, Tellier, C, Dion, M, Leroux, C, Roncza, J, Czjzek, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Semi-Rational Approach for Converting a Gh1 Beta-Glycosidase Into a Beta-Transglycosidase.
Protein Eng.Des.Sel., 27, 2014
|
|
6TPQ
 
 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6LEB
 
 | | Staphylococcus aureus surface protein SdrC mutant-P366H | | Descriptor: | GLYCEROL, MAGNESIUM ION, Ser-Asp rich fibrinogen-binding, ... | | Authors: | Hang, T, Zhang, M, Wang, J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the intermolecular interaction of the adhesin SdrC in the pathogenicity of Staphylococcus aureus.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3ZQE
 
 | | PrgI-SipD from Salmonella typhimurium in complex with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, PROTEIN PRGI, ... | | Authors: | Lunelli, M, Kolbe, M. | | Deposit date: | 2011-06-09 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Prgi-Sipd: Insight Into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Plos Pathog., 7, 2011
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
6TS4
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
259D
 
 | |
7WTD
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
1H4M
 
 | | Sulfurtransferase from Azotobacter vinelandii in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PUTATIVE THIOSULFATE SULFURTRANSFERASE | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
7WTE
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 2 | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
6TS7
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-(1,2,3,4-tetrahydroisoquinolin-7-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
1H73
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH THREONINE | | Descriptor: | HOMOSERINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THREONINE | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
7WTA
 
 | | Cryo-EM structure of human pyruvate carboxylase in apo state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
3ZYV
 
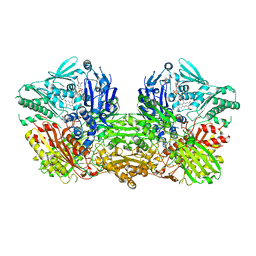 | | Crystal structure of the mouse liver Aldehyde Oxidase 3 (mAOX3) | | Descriptor: | AOX3, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Trincao, J, Coelho, C, Mahro, M, Rodrigues, D, Terao, M, Garattini, E, Leimkuehler, S, Romao, M.J. | | Deposit date: | 2011-08-27 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | The First Mammalian Aldehyde Oxidase Crystal Structure: Insights Into Substrate Specificity.
J.Biol.Chem., 287, 2012
|
|
7WTC
 
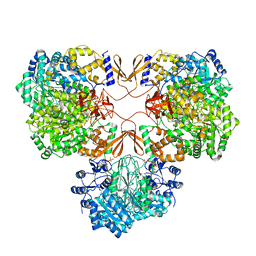 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the ground state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTB
 
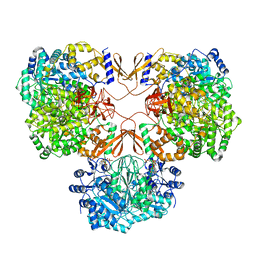 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
3ZKT
 
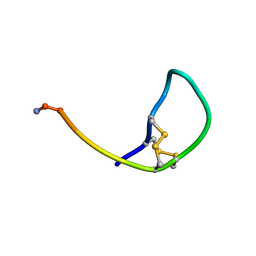 | | SOLUTION STRUCTURE OF THE SOMATOSTATIN SST3 RECEPTOR ANTAGONIST TAU- CONOTOXIN CnVA | | Descriptor: | TAU-CNVA | | Authors: | Petrel, C, Hocking, H.G, Reynaud, M, Favreau, P, Paolini-Bertrand, M, Peigneur, S, Upert, G, Tytgat, J, Gilles, N, Hartley, O, Boelens, R, Stocklin, R, Servent, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, Structural and Pharmacological Characterization of Tau-Cnva, a Conopeptide that Selectively Interacts with Somatostatin Sst3 Receptor.
Biochem.Pharmacol, 85, 2013
|
|
