6A4J
 
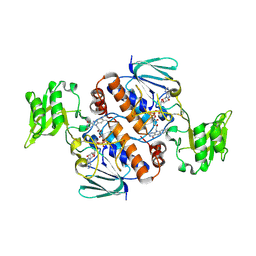 | | Crystal structure of Thioredoxin reductase 2 from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Bose, M, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
6A5P
 
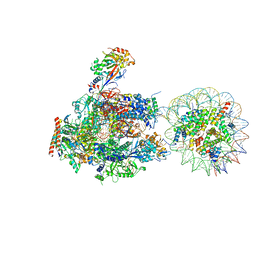 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
5ZVR
 
 | |
5ZWE
 
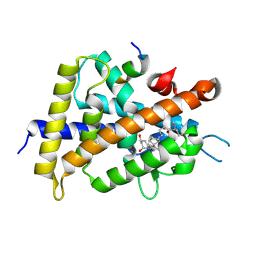 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
6AAC
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mAzZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAR
 
 | | Crystal structure of DAPK1 in complex with purpurin | | Descriptor: | Death-associated protein kinase 1, Purpurin | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic analyses of interactions between death-associated protein kinase 1 and anthraquinones.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6AB1
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oAzZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-azidophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABL
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oBrZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-bromophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
6AFK
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
5ZUJ
 
 | | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA | | Descriptor: | TNF receptor-associated factor 6, peptide 170-184 from TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Huang, W.C, Maestre-Reyna, M, Hsiao, T.C, Tsai, M.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Chembiochem, 20, 2019
|
|
5ZUY
 
 | |
6A5U
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABM
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pTmdZLys | | Descriptor: | (2S)-2-azanyl-6-[[4-[3-(trifluoromethyl)-1,2-diazirin-3-yl]phenyl]methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AJA
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADPH, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6ADE
 
 | | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dilawari, R, Singh, P.K, Raje, M, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution
To Be Published
|
|
6AK1
 
 | | Crystal structure of DmoA from Hyphomicrobium sulfonivorans | | Descriptor: | Dimethyl-sulfide monooxygenase | | Authors: | Cao, H.Y, Wang, P, Peng, M, Li, C.Y. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
6AKO
 
 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6AB6
 
 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AA8
 
 | |
6A5I
 
 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
