4TQ6
 
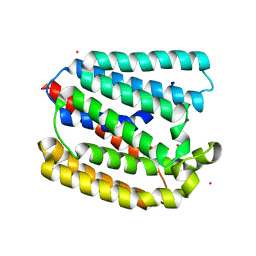 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | Descriptor: | CADMIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0678 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TT4
 
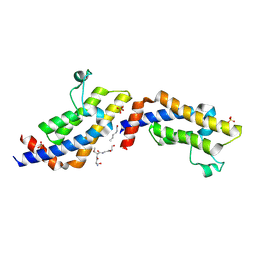 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTU
 
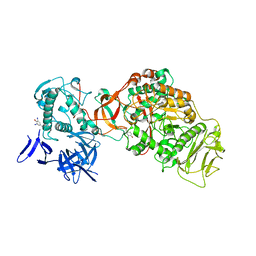 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with isomaltotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
7UAX
 
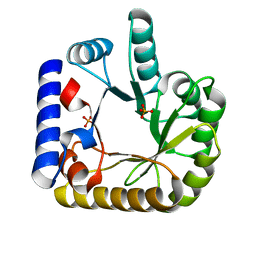 | | The crystal structure of the K36A/K38A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
8SSI
 
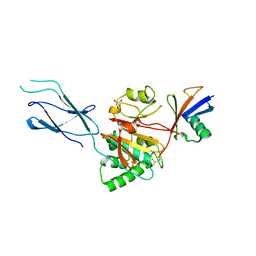 | |
7U9H
 
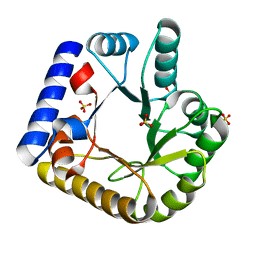 | | Crystal Structure of Escherichia coli apo Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAU
 
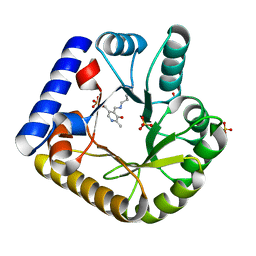 | | The crystal structure of the K137A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
8SDX
 
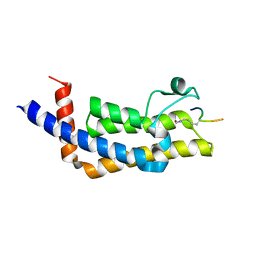 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 5 with Serine 1 mutation to Cysteine | | Descriptor: | ATPase family AAA domain-containing protein 2B, SULFATE ION, histone H4S1CK5ac | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-04-07 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
7UBP
 
 | | The crystal structure of the K36A/K137A double mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
4TQ5
 
 | | Structure of a UbiA homolog from Archaeoglobus fulgidus | | Descriptor: | octyl beta-D-glucopyranoside, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2023 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
7U9C
 
 | | Crystal Structure of the wild type Escherichia coli Pyridoxal 5'-phosphate homeostasis protein (YGGS) | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Safo, M.K, Musayev, F.N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UAT
 
 | | The crystal structure of the K36A mutant of E. coli YGGS in complex with PLP | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB8
 
 | | The crystal structure of the K38A/K137A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | 1,4-BUTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UBQ
 
 | | The crystal structure of the wild-type of E. coli YGGS in complex with PNP | | Descriptor: | PYRIDOXINE-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UB4
 
 | | The crystal structure of the K36A/K38A/K233A/K234A quadruple mutant of E. coli YGGS in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate homeostasis protein | | Authors: | Donkor, A.K, Ghatge, M.S, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of the Escherichia coli pyridoxal 5'-phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability.
Protein Sci., 31, 2022
|
|
7UD8
 
 | | Crystal structure of carbon monoxy Hemoglobin in complex with 5HMF at 1.8 Angstrom | | Descriptor: | (5-methylfuran-2-yl)methanol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.S. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Biomolecules, 12, 2022
|
|
8T5B
 
 | |
4RNL
 
 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
8T5A
 
 | |
6HEE
 
 | | Crystal structure of Extracellular Domain 1 (ECD1) of FtsX from S. pneumonie in complex with undecyl-maltoside | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsX, SULFATE ION, ... | | Authors: | Martinez-Caballero, S, Alcorlo-Pages, M, Hermoso, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
Mbio, 10, 2019
|
|
8T52
 
 | |
6HFX
 
 | |
4RSO
 
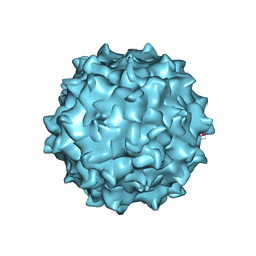 | | The structure of the neurotropic AAVrh.8 viral vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Halder, S, Van Vliet, K, Smith, J.K, McKenna, R, Wilson, J.M, Agbandje-McKenna, M. | | Deposit date: | 2014-11-10 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of neurotropic adeno-associated virus AAVrh.8.
J.Struct.Biol., 192, 2015
|
|
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
5MBY
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, HYDROXIDE ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
