1FOZ
 
 | | STRUCTURE OF CYCLIC PEPTIDE INHIBITORS OF MAMMALIAN RIBONUCLEOTIDE REDUCTASE | | Descriptor: | SYNTHETIC CYCLIC PEPTIDE | | Authors: | Pellegrini, M, Liehr, S, Fisher, A.L, Cooperman, B.S, Mierke, D.F. | | Deposit date: | 2000-08-29 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-based optimization of peptide inhibitors of mammalian ribonucleotide reductase.
Biochemistry, 39, 2000
|
|
1FOA
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
7V63
 
 | | Structure of dimeric uPAR at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2021-08-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
8W34
 
 | | HIV-1 intasome core assembled with wild-type integrase, 1F | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8VHC
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
1F8U
 
 | | CRYSTAL STRUCTURE OF MUTANT E202Q OF HUMAN ACETYLCHOLINESTERASE COMPLEXED WITH GREEN MAMBA VENOM PEPTIDE FASCICULIN-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN II | | Authors: | Kryger, G, Harel, M, Shafferman, A, Silman, I, Sussman, J.L. | | Deposit date: | 2000-07-05 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FAA
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (LONG FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1F9K
 
 | | WINGED BEAN ACIDIC LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE | | Descriptor: | ACIDIC LECTIN, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Carbohydrate specificity and salt-bridge mediated conformational change in acidic winged bean agglutinin.
J.Mol.Biol., 302, 2000
|
|
8W09
 
 | | HIV-1 wild-type intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
1FC0
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1FFX
 
 | | TUBULIN:STATHMIN-LIKE DOMAIN COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PROTEIN (STATHMIN-LIKE DOMAIN OF RB3), ... | | Authors: | Gigant, B, Martin-Barbey, C, Knossow, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | The 4 A X-ray structure of a tubulin:stathmin-like domain complex.
Cell(Cambridge,Mass.), 102, 2000
|
|
6H1N
 
 | |
6H64
 
 | | Crystal structure of the CRD-SAT | | Descriptor: | Galectin-3, SULFATE ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Charron, C, Kriznik, A, Yelehe-Okouma, M, Jouzeau, J.-Y, Reboul, P. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRD SAT Generated by pCARGHO: A New Efficient Lectin-Based Affinity Tag Method for Safe, Simple, and Low-Cost Protein Purification.
Biotechnol J, 14, 2019
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
7VB3
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aliphatic (R)-hydroxynitrile lyase, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
8V04
 
 | | High resolution TMPRSS2 structure following acylation by nafamostat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Levon, H, Arrowsmith, C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High resolution TMPRSS2 structure following acylation by nafamostat
To Be Published
|
|
3E7F
 
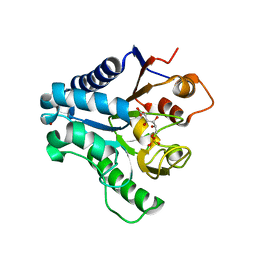 | | Crystal structure of 6-phosphogluconolactonase from Trypanosoma brucei complexed with 6-phosphogluconic acid | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconolactonase, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-18 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
7VK2
 
 | | Crystal Structure of SARS-CoV-2 Mpro at 2.0 A resolution -9 | | Descriptor: | 3C-like proteinase | | Authors: | DeMirci, H, Gul, M. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography
Crystals, 11, 2021
|
|
7VB6
 
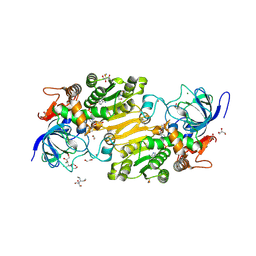 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
1F9U
 
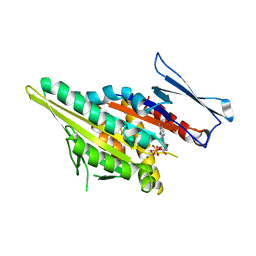 | | CRYSTAL STRUCTURES OF MUTANTS REVEAL A SIGNALLING PATHWAY FOR ACTIVATION OF THE KINESIN MOTOR ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KAR3, MAGNESIUM ION | | Authors: | Yun, M, Zhang, X, Park, C.G, Park, H.W, Endow, S.A. | | Deposit date: | 2000-07-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural pathway for activation of the kinesin motor ATPase.
EMBO J., 20, 2001
|
|
7VB5
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissimum complexed with acetone cyanohydrin | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXY-2-METHYLPROPANENITRILE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
1FP9
 
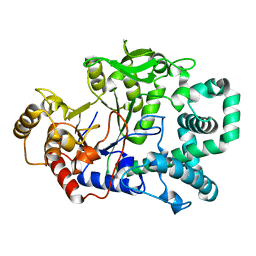 | |
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
