3Q8E
 
 | | Crystal structure of Protective Antigen W346F (pH 8.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3DTO
 
 | | Crystal structure of the metal-dependent HD domain-containing hydrolase BH2835 from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR130. | | Descriptor: | BH2835 protein | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owen, L.A, Wang, D, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the metal-dependent HD domain-containing hydrolase BH2835 from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR130.
To be Published
|
|
1LU4
 
 | | 1.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED MYCOBACTERIUM TUBERCULOSIS DISULFIDE OXIDOREDUCTASE HOMOLOGOUS TO E. COLI DSBE: IMPLICATIONS FOR FUNCTIONS | | Descriptor: | SOLUBLE SECRETED ANTIGEN MPT53 | | Authors: | Goulding, C.W, Apostol, M.I, Gleiter, S, Parseghian, A, Bardwell, J, Gennaro, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Gram-positive DsbE Proteins Function Differently from Gram-negative DsbE Homologs: A STRUCTURE TO FUNCTION ANALYSIS OF DsbE FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 279, 2004
|
|
3QNF
 
 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6KAI
 
 | | Crosslinked alpha(Ni)-beta(Fe) human hemoglobin A in the T quaternary structure at 95 K: Light | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-23 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4KLK
 
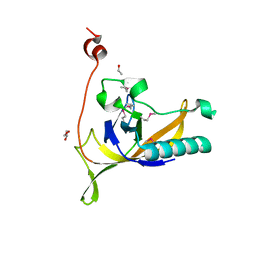 | | Phage-related protein DUF2815 from Enterococcus faecalis | | Descriptor: | ETHANOL, GLYCEROL, Phage-related protein DUF2815 | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phage-related protein DUF2815 from Enterococcus faecalis
To be Published
|
|
6KAS
 
 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 95 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Q6T
 
 | | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5 | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5
To be Published
|
|
3QPB
 
 | | Crystal Structure of Streptococcus Pyogenes Uridine Phosphorylase Reveals a Subclass of the NP-I Superfamily | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, URACIL, Uridine phosphorylase | | Authors: | Tran, T.H, Christoffersen, S, Parker, W.B, Piskur, J, Serra, I, Terreni, M, Ealick, S.E. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Streptococcus pyogenes Uridine Phosphorylase Reveals a Distinct Subfamily of Nucleoside Phosphorylases.
Biochemistry, 50, 2011
|
|
5REA
 
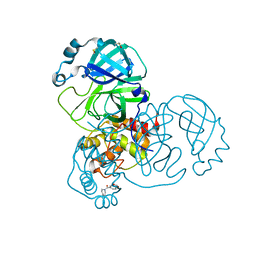 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z31432226 | | Descriptor: | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4Q1S
 
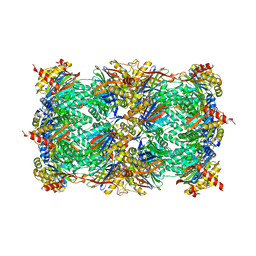 | | Yeast 20S proteasome in Complex with Kendomycin | | Descriptor: | (5R,6R,7S,8R,9R,12S,13E,16S,18S,19R,20aR)-4,7,19-trihydroxy-2,6,8,12,14,16,18-heptamethyl-6,7,8,9,10,11,12,15,16,17,18,19,20,20a-tetradecahydro-1,19:5,9-diepoxybenzo[18]annulen-3(5H)-one, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Beck, P, Heinemeyer, W, Spaeth, A, Elnakady, Y, Mueller, R, Groll, M. | | Deposit date: | 2014-04-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of the natural product kendomycin and the 20S proteasome.
J.Mol.Biol., 426, 2014
|
|
4Q3E
 
 | | PylD cocrystallized with L-Ornithine-Nd-D-lysine and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5RES
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102281 | | Descriptor: | 1-{4-[(2-fluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4PW6
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
6K9L
 
 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
5S2O
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z645232558 | | Descriptor: | Non-structural protein 3, [1-(pyrimidin-2-yl)piperidin-4-yl]methanol | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.091 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4PWF
 
 | | Crystal structure of V30M mutant human transthyretin complexed with ferulic acid phenethyl ester | | Descriptor: | 2-phenylethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2014-03-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitory Activities of Propolis and Its Promising Component, Caffeic Acid Phenethyl Ester, against Amyloidogenesis of Human Transthyretin
J.Med.Chem., 57, 2014
|
|
5S3Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0140 | | Descriptor: | (3R,4S)-4-(3-methoxyphenyl)oxan-3-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S35
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z68404778 | | Descriptor: | Non-structural protein 3, ~{N}-(4-phenylazanylphenyl)ethanamide | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3DXH
 
 | | Ribonuclease A uridine 5' diphosphate complex | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-DIPHOSPHATE | | Authors: | Tsirkone, V.G, Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Kontou, M. | | Deposit date: | 2008-07-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibitor design to Ribonuclease A: The binding of two 5'phosphate uridine analogues
Acta Crystallogr.,Sect.F, 2009
|
|
2N7T
 
 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
4L4M
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
5S3L
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z54628578 | | Descriptor: | N-methylpyrimidin-2-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.091 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3QTD
 
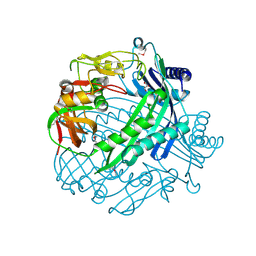 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
5S42
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023833 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
