6H57
 
 | | Crystal structure of S. cerevisiae DEAH-box RNA helicase Dhr1, essential for small ribosomal subunit biogenesis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Roychowdhury, A, Graille, M. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis.
Nucleic Acids Res., 47, 2019
|
|
4PB5
 
 | | D-threo-3-hydroxyaspartate dehydratase H351A mutant complexed with L-erythro-3-hydroxyaspartate | | Descriptor: | (3R)-3-hydroxy-L-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3PS8
 
 | | Crystal structure of L68V mutant of human cystatin C | | Descriptor: | ACETATE ION, Cystatin-C, DI(HYDROXYETHYL)ETHER | | Authors: | Orlikowska, M, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | Deposit date: | 2010-12-01 | | Release date: | 2011-12-21 | | Last modified: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of L68V mutant of human cystatin C
To be Published
|
|
4J58
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Colliandre, L, Gelin, M, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1HM7
 
 | |
6SSU
 
 | |
6F6P
 
 | |
1HN0
 
 | |
2C43
 
 | | STRUCTURE OF AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE- PHOSPHOPANTETHEINYL TRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Oppermann, U. | | Deposit date: | 2005-10-14 | | Release date: | 2005-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
6GP5
 
 | | Beta-1,4-galactanase from Bacteroides thetaiotaomicron | | Descriptor: | Arabinogalactan endo-beta-1,4-galactanase | | Authors: | Hekelaar, J, Boger, M, Leeuwen van, S.S, Lammerts van Bueren, A, Dijkhuizen, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and functional characterization of a family GH53 beta-1,4-galactanase from Bacteroides thetaiotaomicron that facilitates degradation of prebiotic galactooligosaccharides.
J. Struct. Biol., 205, 2019
|
|
3PVR
 
 | |
1HO2
 
 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN MICELLES | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-08 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
6F6Y
 
 | |
6H6F
 
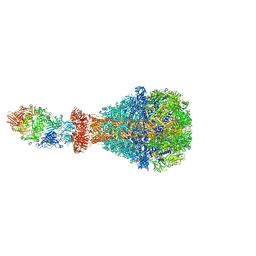 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
4P1G
 
 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
2MXO
 
 | |
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
4P4L
 
 | | Crystal Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, Shikimate 5-dehydrogenase AroE (5-dehydroshikimate reductase) | | Authors: | Lalgondar, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-03-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase
To Be Published
|
|
6SLI
 
 | | Structure of the RagAB peptide transporter | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALA-SER-THR-THR-GLY-GLY-ASN-SER-GLN-ARG-GLY-SER-GLY, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-19 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6F9H
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | Descriptor: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
4P4G
 
 | | Crystal Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, BROMIDE ION, SULFATE ION, ... | | Authors: | Lalgondar, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-03-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Dehydrogenase
To Be Published
|
|
2N71
 
 | | NMR structure of CmPI-II, a serin protease inhibitor isolated from mollusk Cenchitis muricatus | | Descriptor: | Protease inhibitor 2 | | Authors: | Cabrera-Munoz, A, Rojas, L, Alonso del Rivero Antigua, M, Pires, J. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of CmPI-II, a non-classical Kazal protease inhibitor: Understanding its conformational dynamics and subtilisin A inhibition.
J.Struct.Biol., 206, 2019
|
|
6SP8
 
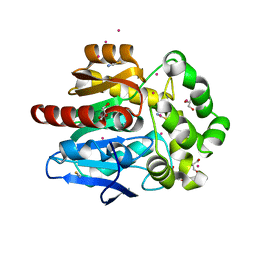 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6H26
 
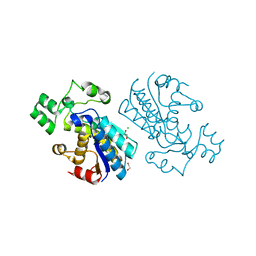 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
1HIT
 
 | |
