4U9N
 
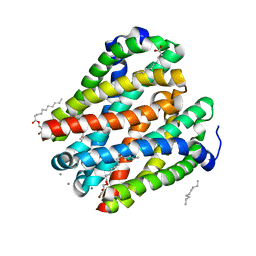 | | Structure of a membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MANGANESE (II) ION, Magnesium transporter MgtE, ... | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4UB8
 
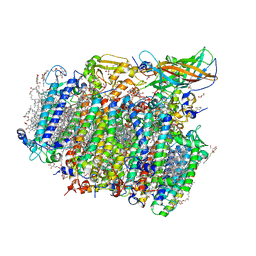 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
6WNZ
 
 | |
4UDS
 
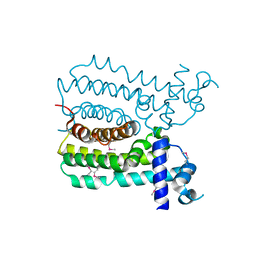 | | Crystal structure of MbdR regulator from Azoarcus sp. CIB | | Descriptor: | MBDR REGULATOR | | Authors: | Liu, H, Liu, H.X, MacMahon, S.A, Juarez, J.F, Zamarro, M.T, Eberlein, C, Boll, M, Carmona, M, Diaz, E, Naismith, J.H. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unraveling the Specific Regulation of the Mbd Central Pathway for the Anaerobic Degradation of 3-Methylbenzoate.
J.Biol.Chem., 290, 2015
|
|
6QXN
 
 | | Crystal structure of the CCA-adding enzyme of a psychrophilic organism in complex with CTP | | Descriptor: | ACETATE ION, CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | de Wijn, R, Rollet, K, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus
Comput Struct Biotechnol J, 2021
|
|
4UPB
 
 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
7SF2
 
 | | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-02 | | Release date: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus
To Be Published
|
|
4UP5
 
 | | Crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a chemical fragment | | Descriptor: | 6-methoxy-1,3-benzothiazol-2-amine, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
4UNE
 
 | | Human insulin B26Phe mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7S0F
 
 | | Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi protein | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Paknejad, N, Alegre, K.O, Su, M, Lou, J.S, Huang, J, Jordan, K.D, Eng, E.T, Meyerson, J.R, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and mechanism of activation of two different families of G proteins by the same GPCR.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7S0G
 
 | | Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi/s chimera protein | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Paknejad, N, Alegre, K.O, Su, M, Lou, J.S, Huang, J, Jordan, K.D, Eng, E.T, Meyerson, J.R, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis and mechanism of activation of two different families of G proteins by the same GPCR.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6R2E
 
 | | Crystal structure of the human thymidylate synthase (hTS) interface variant Q62R | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Biomolecules, 9, 2019
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XE1
 
 | |
6X9Y
 
 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
7SJZ
 
 | | Crystal structure of aS162A mutant of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Ogutu, R.A.M.I, Holz, C.R, Bennett, B, St Maurice, M. | | Deposit date: | 2021-10-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Examination of the Catalytic Role of the Axial Cystine Ligand in the Co-Type Nitrile Hydratase from Pseudonocardia thermophila JCM 3095
Catalysts, 11, 2021
|
|
4UP4
 
 | | Structure of the recombinant lectin PVL from Psathyrella velutina in complex with GlcNAcb-D-1,3Galactoside | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Audfray, A, Beljoudi, M, Hurbin, A, Varrot, A, Breiman, A, Busser, B, Lependu, J, Coll, J.L, Imberty, A. | | Deposit date: | 2014-06-12 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Recombinant Fungal Lectin for Labeling Truncated Glycans on Human Cancer Cells.
Plos One, 10, 2015
|
|
6XKV
 
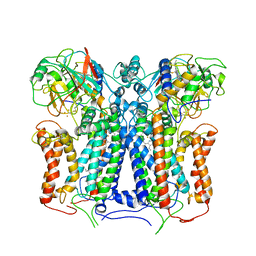 | | R. capsulatus cyt bc1 with both FeS proteins in b position (CIII2 b-b) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
7RY5
 
 | | Cellular Retinoic Acid Binding Protein II with Bound Inhibitor 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid | | Descriptor: | 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Kimmel, B.R, Mrksich, M. | | Deposit date: | 2021-08-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Enzyme-Inhibitor Reaction Using Cellular Retinoic Acid Binding Protein II for One-Pot Megamolecule Assembly.
Chemistry, 27, 2021
|
|
6X7K
 
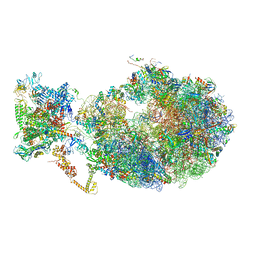 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-30 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
4UNI
 
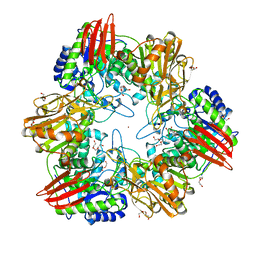 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UB6
 
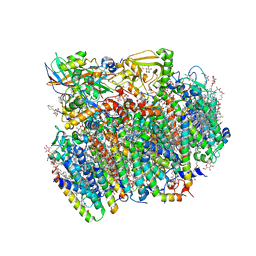 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
6XE0
 
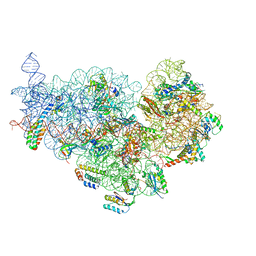 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
7S56
 
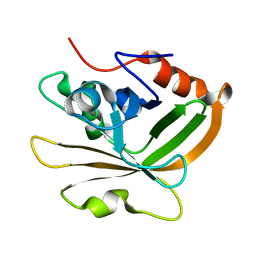 | | Sortase A from Streptococcus agalactiae, residues 79-247 | | Descriptor: | Class A sortase | | Authors: | Gao, M, Kodama, H.M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
4UII
 
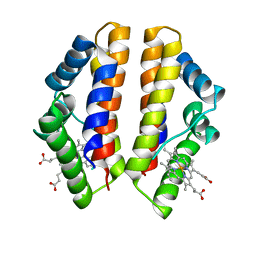 | | Crystal structure of the Azotobacter vinelandii globin-coupled oxygen sensor in the aquo-met form | | Descriptor: | GGDEF DOMAIN PROTEIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Germani, F, De Schutter, A, Pesce, A, Berghmans, H, Van Hauwaert, M.-L, Cuypers, B, Bruno, S, Mozzarelli, A, Moens, L, Van Doorslaer, S, Bolognesi, M, Nardini, M, Dewilde, S. | | Deposit date: | 2015-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.827 Å) | | Cite: | Azotobacter Vinelandii Globin-Coupled Oxygen Sensor is a Diguanylate Cyclase with a Biphasic Oxygen Dissociation
To be Published
|
|
