6Z65
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-4-azanyl-butanamide | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
6Z13
 
 | | VEGF-A 13:107 crystallized with 3C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
6Z64
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-(3-azanylpropyl)amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
4LQ5
 
 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | rystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium LT2 in complex with effector ligand, O-acetylserine at 2.8A
TO BE PUBLISHED
|
|
6ZGD
 
 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6Z3F
 
 | | VEGF-A 13:107 crystallized with 2C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
6ZGJ
 
 | | GLIC pentameric ligand-gated ion channel, pH 5 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
3SCK
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3SD8
 
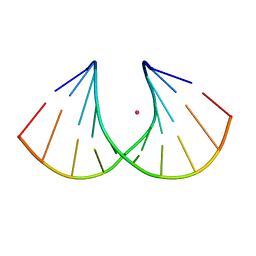 | | Crystal structure of Ara-FHNA decamer DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(F4H)P*AP*CP*GP*C)-3', STRONTIUM ION | | Authors: | Egli, M, Pallan, P.S. | | Deposit date: | 2011-06-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Origins of Opposite Effects on Stability by Axial and Equatorial 3'-Fluoro Modifications of Hexitol Nucleic Acid (HNA)
To be Published
|
|
3SN6
 
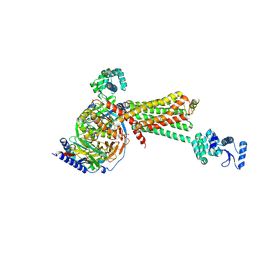 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
6Z6E
 
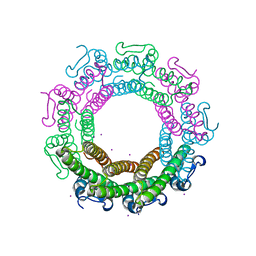 | | Crystal structure of the HK97 bacteriophage small terminase | | Descriptor: | IODIDE ION, Terminase small subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
3RZS
 
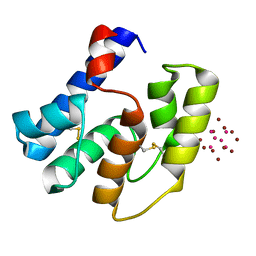 | | Apis mellifera OBP14 in complex with Ta6Br14 | | Descriptor: | HEXATANTALUM DODECABROMIDE, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-12 | | Release date: | 2011-11-30 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
4M9M
 
 | |
4LXS
 
 | | Structure of the Toll - Spatzle complex, a molecular hub in Drosophila development and innate immunity (glycosylated form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stelter, M, Parthier, C, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2013-07-30 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SM1
 
 | |
6Z6D
 
 | | Crystal structure of the HK97 bacteriophage large terminase | | Descriptor: | BROMIDE ION, Terminase large subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4MA7
 
 | | Crystal structure of mouse prion protein complexed with Promazine | | Descriptor: | Major prion protein, POM1 heavy chain, POM1 light chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
6ZBR
 
 | | VEGF-A 13:107 crystallized with 4C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chains: P, PHOSPHATE ION, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
4M0I
 
 | |
6Z2S
 
 | | Crystal structure of adhibin analogue-bound myosin-2 | | Descriptor: | 3,6-dibromo-1-(hydroxymethyl)carbazole, ACETATE ION, ADP METAVANADATE, ... | | Authors: | Ewert, W, Knoelker, H.-J, Tsiavaliaris, G, Preller, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The synthetic carbazole adhibin acts as selective myosin class-IX inhibitor in lung cancer cells
To Be Published
|
|
6ZCD
 
 | | VEGF-A 13:107 crystallized with 1C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Derived from V114 peptide, PHOSPHATE ION, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
4MDY
 
 | |
4M3E
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3S5M
 
 | | Crystal structures of falcilysin, a M16 metalloprotease from the malaria parasite Plasmodium falciparum | | Descriptor: | Falcilysin, MAGNESIUM ION, ZINC ION | | Authors: | Morgunova, E, Ponpuak, M, Istvan, E, Popov, A, Goldberg, D, Eneqvist, T. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of falcilysin, a M16 metalloprotease from
the malaria parasite Plasmodium falciparum
To be Published
|
|
