8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
6PSA
 
 | | PIE12 D-PEPTIDE AGAINST HIV ENTRY (IN COMPLEX WITH IQN17 Q577R RESISTANCE MUTANT) | | Descriptor: | CHLORIDE ION, IQN17, PIE12 D-peptide | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Weinstock, M. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of resistance to a potent D-peptide HIV entry inhibitor.
Retrovirology, 16, 2019
|
|
6PXA
 
 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | Descriptor: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | Authors: | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid
To Be Published
|
|
4MH7
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1896 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-butyl-2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-methylpyrimidine-5-carboxamide, ... | | Authors: | Zhang, W, McIver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, M.J, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, DiPaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
7CYI
 
 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua | | Descriptor: | Alcohol dehydrogenase 1, ZINC ION | | Authors: | Feng, X, Fan, S, Lv, G, Li, M, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua
To Be Published
|
|
1RZF
 
 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6NPO
 
 | | Crystal structure of oligopeptide ABC transporter from Bacillus anthracis str. Ames (substrate-binding domain) | | Descriptor: | Oligopeptide ABC transporter, oligopeptide-binding protein, Unknown peptide ligand, ... | | Authors: | Michalska, K, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oligopeptide ABC transporter from
Bacillus anthracis str. Ames (substrate-binding domain)
To Be Published
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
2JDY
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-b-D-mannoyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-D-mannopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
5NQ8
 
 | | Crystal structure of laccases from Pycnoporus sanguineus, izoform II | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, COPPER (II) ION, Laccase, ... | | Authors: | Orlikowska, M, de J.Rostro-Alanis, M, Bujacz, A, Hernandez-Luna, C, Rubio, R, Parra, R, Bujacz, G. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus.
Int. J. Biol. Macromol., 107, 2018
|
|
6NPN
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor | | Descriptor: | (7R)-7-methyl-2-[(3-oxo-2,3-dihydro-1H-indazol-6-yl)amino]-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, dos Reis, C.V, Takarada, J.E, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor
To Be Published
|
|
6Q2E
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
6Q2P
 
 | | Crystal structure of mouse viperin bound to cytidine triphosphate and S-adenosylhomocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Structural Basis of the Substrate Selectivity of Viperin.
Biochemistry, 59, 2020
|
|
7DHS
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
6Q94
 
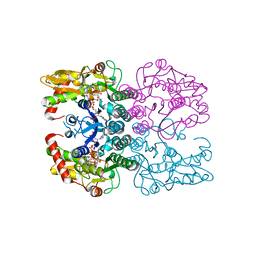 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
2PBH
 
 | |
4ALT
 
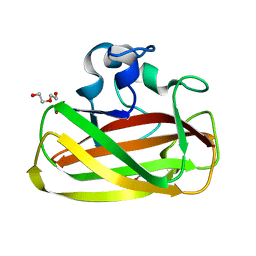 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
6NV2
 
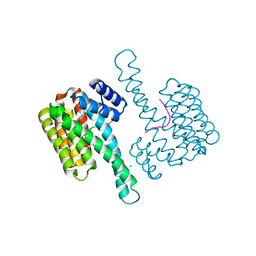 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
5N67
 
 | |
6QA4
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-pyridin-2-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
4LZR
 
 | | Crystal Structure of BRD4(1) bound to Colchicine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O, Huegle, M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5DE0
 
 | | Dye-decolorizing protein from V. cholerae | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Uchida, T, Sasaki, M, Tanaka, Y, Yao, M. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A Dye-Decolorizing Peroxidase from Vibrio cholerae.
Biochemistry, 54, 2015
|
|
1YMU
 
 | |
6QBE
 
 | | Crystal structure of non-toxic HaNLP3 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nep1-like protein, PHOSPHATE ION, ... | | Authors: | Lenarcic, T, Podobnik, M, Anderluh, G. | | Deposit date: | 2018-12-21 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for functional diversity among microbial Nep1-like proteins.
Plos Pathog., 15, 2019
|
|
1YMV
 
 | |
