1HBW
 
 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | Descriptor: | REGULATORY PROTEIN GAL4 | | Authors: | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
2Q3H
 
 | | The crystal structure of RhouA in the GDP-bound state. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras homolog gene family, ... | | Authors: | Gileadi, C, Yang, X, Papagrigoriou, E, Elkins, J, Zhao, Y, Bray, J, Gileadi, O, Umeano, C, Ugochukwu, E, Uppenberg, J, Bunkoczi, G, von Delft, F, Pike, A.C.W, Phillips, C, Savitsky, P, Fedorov, O, Edwards, A, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of RhouA in the GDP-bound state.
To be Published
|
|
3BFA
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
2PQ2
 
 | | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution | | Descriptor: | CALCIUM ION, GALAG peptide, NITRATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, A.K, Singh, N, Sharma, S, Sinha, M, Somvanshi, R.K, Kaur, P, Dey, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution
To be Published
|
|
9AXU
 
 | | Non-translating S. pombe ribosome large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Gluc, M, Gemin, O, Purdy, M, Mattei, S, Jomaa, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Ribosomes hibernate on mitochondria during cellular stress.
Nat Commun, 15, 2024
|
|
1QFO
 
 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Vinson, M, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
1GVC
 
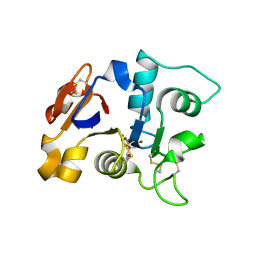 | | 18kDa N-II domain fragment of duck ovotransferrin + NTA | | Descriptor: | CARBONATE ION, FE (III) ION, NITRILOTRIACETIC ACID, ... | | Authors: | Kuser, P, Hall, D.R, Haw, M.L, Neu, M, Lindley, P.F. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism of Iron Uptake by Transferrins: The X-Ray Structures of the 18 kDa Nii Domain Fragment of Duck Ovotransferrin and its Nitrilotriacetate Complex
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2PQE
 
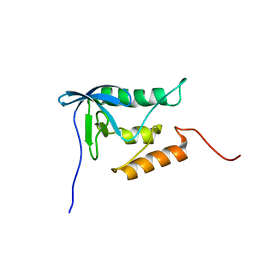 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
9EM1
 
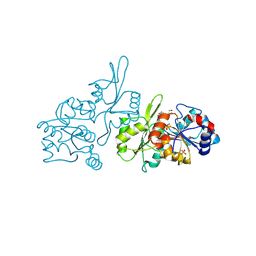 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
1QFX
 
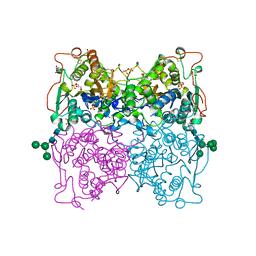 | | PH 2.5 ACID PHOSPHATASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PROTEIN (PH 2.5 ACID PHOSPHATASE), ... | | Authors: | Kostrewa, D, Wyss, M, D'Arcy, A, Van Loon, A.P.G.M. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2. 4 A resolution.
J.Mol.Biol., 288, 1999
|
|
6ZJZ
 
 | | Discovery of M5049: a novel selective TLR7/8 inhibitor for treatment of autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3~{R},5~{S})-3-azanyl-5-(trifluoromethyl)piperidin-1-yl]quinoline-8-carbonitrile, FORMIC ACID, ... | | Authors: | Musil, D, Lehman, M, Strauss, J. | | Deposit date: | 2020-06-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Discovery of M5049: A Novel Selective Toll-Like Receptor 7/8 Inhibitor for Treatment of Autoimmunity.
J.Pharmacol.Exp.Ther., 376, 2021
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2PSQ
 
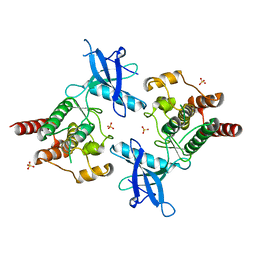 | |
1QI2
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH 2',4'-DINITROPHENYL 2-DEOXY-2-FLUORO-B-D-CELLOTRIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Varrot, A, Schulein, M, Davies, G.J. | | Deposit date: | 1999-06-02 | | Release date: | 2000-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into ligand-induced conformational change in Cel5A from Bacillus agaradhaerens revealed by a catalytically active crystal form.
J.Mol.Biol., 297, 2000
|
|
2PVY
 
 | |
1GQ1
 
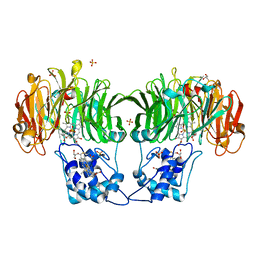 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
3B7T
 
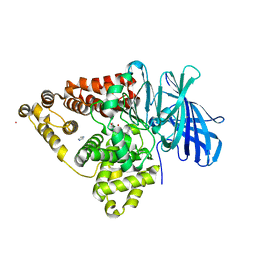 | | [E296Q]LTA4H in complex with Arg-Ala-Arg substrate | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, RAR peptide, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3ZQ6
 
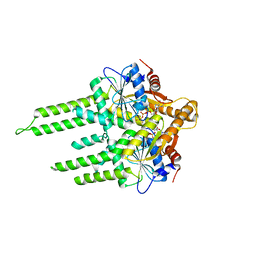 | | ADP-ALF4 COMPLEX OF M. THERM. TRC40 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sherrill, J, Mariappan, M, Dominik, P, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | A Conserved Archaeal Pathway for Tail-Anchored Membrane Protein Insertion.
Traffic, 12, 2011
|
|
3BAP
 
 | |
1QMO
 
 | | Structure of FRIL, a legume lectin that delays hematopoietic progenitor maturation | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, MANNOSE BINDING LECTIN, ... | | Authors: | Hamelryck, T.W, Moore, J.G, Chrispeels, M, Loris, R, Wyns, L. | | Deposit date: | 1999-10-04 | | Release date: | 1999-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Role of Weak Protein-Protein Interactions in Multivalent Lectin-Carbohydrate Binding: Crystal Structure of Cross-Linked Fril
J.Mol.Biol., 299, 2000
|
|
2PYZ
 
 | | Crystal structure of the complex of proteinase K with auramine at 1.8A resolution | | Descriptor: | 4,4'-(AMINOMETHYLENE)BIS(N,N-DIMETHYLANILINE), CALCIUM ION, NITRATE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex of Proteinase K with auramine at 1.8A resolution
To be Published
|
|
2PZR
 
 | |
2PMO
 
 | | Crystal structure of PfPK7 in complex with hymenialdisine | | Descriptor: | 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, Ser/Thr protein kinase | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
3BAV
 
 | |
1QR8
 
 | | INHIBITION OF HIV-1 INFECTIVITY BY THE GP41 CORE: ROLE OF A CONSERVED HYDROPHOBIC CAVITY IN MEMBRANE FUSION | | Descriptor: | GP41 ENVELOPE PROTEIN | | Authors: | Ji, H, Shu, W, Burling, F.T, Jiang, S.B, Lu, M. | | Deposit date: | 1999-06-18 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion.
J.Virol., 73, 1999
|
|
