2QG8
 
 | | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Acyl Carrier Protein Synthase PY06285, MAGNESIUM ION | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Wasney, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Brokx, S, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound
TO BE PUBLISHED
|
|
6F17
 
 | | Structure of Mb NMH H64V, V68A mutant resting state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6F19
 
 | | Structure of Mb NMH H64V, V68A mutant complex with EDA incubated at room temperature for 5 min | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
5EUV
 
 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6SE9
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in shallow mode | | Descriptor: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
2A58
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
6ETI
 
 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F67
 
 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with 3,4-Dihydroxybenzophenone | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, [3,4-bis(oxidanyl)phenyl]-phenyl-methanone, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
2QNW
 
 | | Toxoplasma gondii apicoplast-targeted acyl carrier protein | | Descriptor: | Acyl carrier protein, SODIUM ION, SULFATE ION, ... | | Authors: | Lunin, V.V, Wernimont, A, Lew, J, Qiu, W, Lin, L, Hassanali, A, Kozieradzki, I, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxoplasma gondii apicoplast-targeted acyl carrier protein.
To be Published
|
|
3KZZ
 
 | |
5MQ5
 
 | | A protease-resistant N24S Escherichia coli Asparaginase mutant with outstanding stability and enhanced anti-leukaemic activity | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Maggi, M, Mittelman, S.D, Parmentier, J.H, Whitmire, J.M, Merrell, D.S, Scotti, C. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A protease-resistant Escherichia coli asparaginase with outstanding stability and enhanced anti-leukaemic activity in vitro.
Sci Rep, 7, 2017
|
|
6Y5V
 
 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
2QQE
 
 | | Thymidine Kinase from Thermotoga Maritima in complex with Thymidine | | Descriptor: | THYMIDINE, Thymidine kinase, ZINC ION | | Authors: | Segura-Pena, D, Lichter, J, Trani, M, Konrad, M, Lavie, A, Lutz, S. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quaternary structure change as a mechanism for the regulation of thymidine kinase 1-like enzymes.
Structure, 15, 2007
|
|
2A59
 
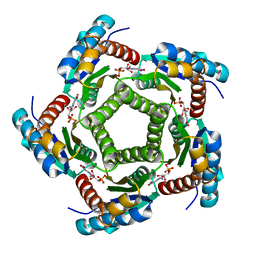 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
6FE6
 
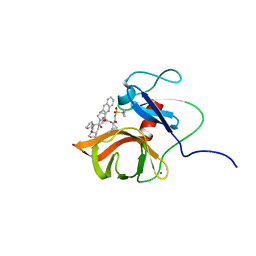 | | Solution structure of a last generation P2-P4 macrocyclic inhibitor | | Descriptor: | (3aR,7S,10S,12R,24aR)-7-cyclopentyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-5,8-dioxo-1,2,3,3a,5,6,7,8,11,12,20,21,22,23,24,24a-hexadecahydro-10H-9,12-methanocyclopenta[18,19][1,10,3,6]dioxadiazacyclononadecino[12,11-b]quinoline-10-carboxamide, Non-structural 3 protease, ZINC ION | | Authors: | Gallo, M, Eliseo, T, Cicero, D.O. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a last generation macrocyclic inhibitor. Hepatitis C virus NS3 protease complex: when S prime region occupancy is not enough to stabilize the protein conformation in the absence of NS4A.
To Be Published
|
|
8FOX
 
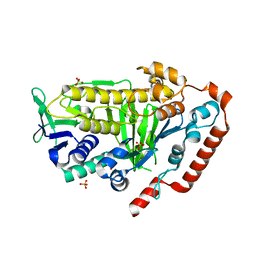 | | AbeH (Tryptophan-5-halogenase) | | Descriptor: | SULFATE ION, Tryptophan 5-halogenase | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
1GX3
 
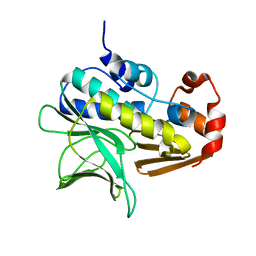 | | M. smegmatis arylamine N-acetyl transferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Mushtaq, A, Kawamura, A, Sinclair, J, Sim, E, Noble, M. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Arylamine N-Acetyltransferase from Mycobacterium Smegmatis-an Enzyme which Inactivates the Anti-Tubercular Drug, Isoniazid
J.Mol.Biol., 318, 2002
|
|
6SEH
 
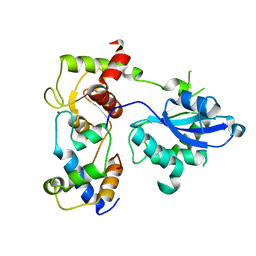 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ZINC ION | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
2A98
 
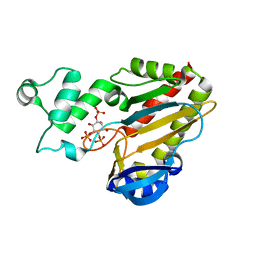 | | Crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate 3-kinase C | | Authors: | Hallberg, B.M, Ogg, D, Ehn, M, Graslund, S, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Stenmark, P, Thorsell, A.-G, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C
To be Published
|
|
2R3A
 
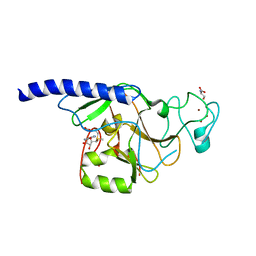 | | Methyltransferase domain of human suppressor of variegation 3-9 homolog 2 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H2, S-ADENOSYLMETHIONINE, SERINE, ... | | Authors: | Lunin, V.V, Wu, H, Zeng, H, Ren, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-29 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
6F68
 
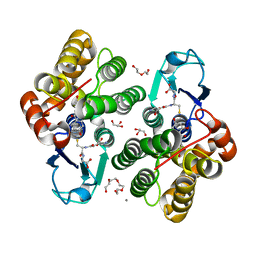 | | Crystal structure glutathione transferase Omega 3S from Trametes versicolor in complex with 2,4,4'-trihydroxybenzophenone | | Descriptor: | (2,4-dihydroxyphenyl)(4-hydroxyphenyl)methanone, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-05 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
8FOV
 
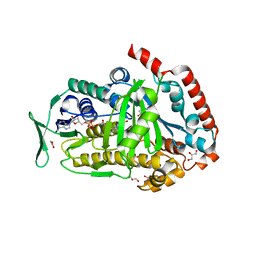 | | AbeH (Tryptophan-5-halogenase) bound to FAD and Cl | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ashaduzzaman, M, Bellizzi, J.J. | | Deposit date: | 2023-01-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
7AAW
 
 | | Thioredoxin Reductase from Bacillus cereus | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shoor, M, Gudim, I, Hersleth, H.-P, Hammerstad, M. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thioredoxin reductase from Bacillus cereus exhibits distinct reduction and NADPH-binding properties.
Febs Open Bio, 11, 2021
|
|
8FLJ
 
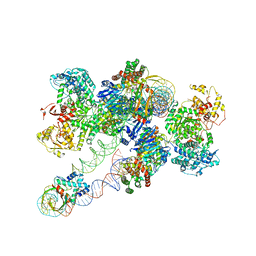 | | Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA | | Descriptor: | CRISPR leader and repeat, anti-sense strand of DNA, CRISPR leader, ... | | Authors: | Santiago-Frangos, A, Henriques, W.S, Wiegand, T, Gauvin, C, Buyukyoruk, M, Neselu, K, Eng, E.T, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5F34
 
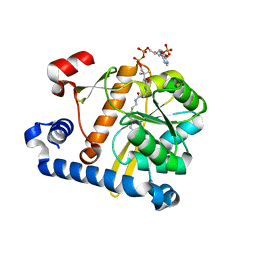 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with S-hexadecyl Coenzyme A - P21 space group | | Descriptor: | Phosphatidylinositol mannoside acyltransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-(2-hexadecylsulfanylethylamino)-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
