7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
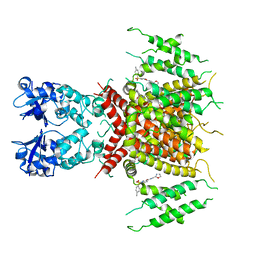 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PV7
 
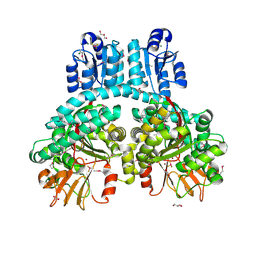 | | Crystal structure of dimeric Porphyromonas gingivalis PorX, a type 9 secretion system response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Nucleic Acids Res., 50, 2022
|
|
7PVA
 
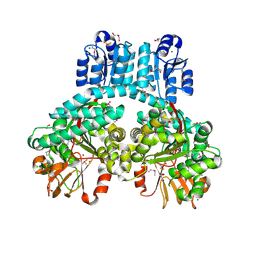 | | 1.9 Angstrom crystal structure of dimeric PorX, co-crystallized in the presence of zinc | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Nucleic Acids Res., 50, 2022
|
|
7PVK
 
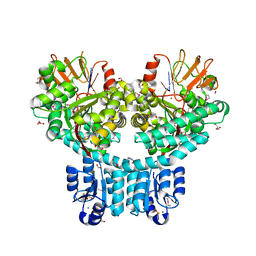 | | X-ray structure of dimeric PorX (T272A mutant), in complex with pGpG. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-04 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors
Nucleic Acids Res., 50, 2022
|
|
7PK4
 
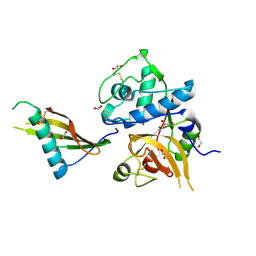 | | Tick salivary cystatin Ricistatin in complex with cathepsin V | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Busa, M, Mares, M. | | Deposit date: | 2021-08-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Protease-bound structure of Ricistatin provides insights into the mechanism of action of tick salivary cystatins in the vertebrate host.
Cell.Mol.Life Sci., 80, 2023
|
|
6FLG
 
 | |
8BXJ
 
 | |
7L6A
 
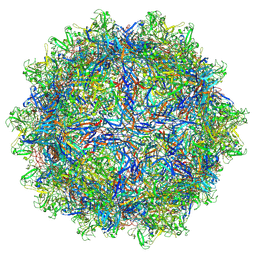 | | The genome-containing AAV12 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L5U
 
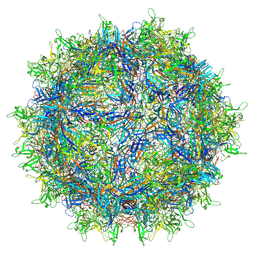 | | The full AAV7 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
8C2Q
 
 | | Silver ion-bound structure of the silver specific chaperone SilF needed for bacterial silver resistance | | Descriptor: | Copper ABC transporter substrate-binding protein, SILVER ION | | Authors: | Monneau, Y.R, Walker, O, Hologne, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | The battle for silver binding: How the interplay between the SilE, SilF, and SilB proteins contributes to the silver efflux pump mechanism.
J.Biol.Chem., 299, 2023
|
|
7L6E
 
 | | The genome-containing AAV11 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6F
 
 | | The empty AAV11 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6H
 
 | | The genome-containing AAV13 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
6LKK
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7L6I
 
 | | The empty AAV13 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7PKU
 
 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
7LH9
 
 | | Crystal structure of BRPF2 PWWP domain in complex with DNA | | Descriptor: | Bromodomain-containing protein 1, DNA | | Authors: | Zhang, M, Lei, M, Qin, S, Dong, A, Yang, A, Li, Y, Loppnau, P, Hughes, T.R, Arrowsmith, C.H, Edwards, A.M, Min, J, Liu, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the BRPF2 PWWP domain in complex with DNA reveals a different binding mode than the HDGF family of PWWP domains.
Biochim Biophys Acta Gene Regul Mech, 1864, 2021
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8JYG
 
 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6LQN
 
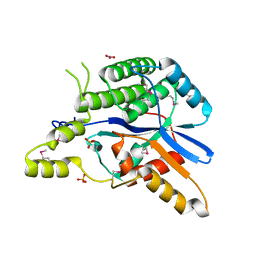 | | EBV tegument protein BBRF2 | | Descriptor: | Cytoplasmic envelopment protein 1, NITRATE ION, SULFATE ION | | Authors: | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.600022 Å) | | Cite: | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
5CLM
 
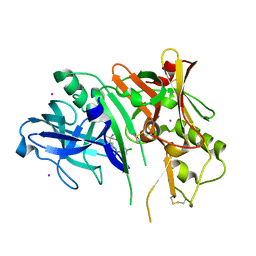 | | 1,4-Oxazine BACE1 inhibitors | | Descriptor: | Beta-secretase 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rombouts, F, Tresadern, G, Delgado, O, Martinez Lamenca, C, Van Gool, M, Garcia-Molina, A, Alonso De Diego, S, Oehlrich, D, Prokopcova, H, Alonso, J.M, Austin, N, Borghys, H, Van Brandt, S, Surkyn, M, De Cleyn, M, Vos, A, Alexander, R, MacDonald, G, Moechars, D, Trabanco, A, Gijsen, H. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 1,4-Oxazine beta-Secretase 1 (BACE1) Inhibitors: From Hit Generation to Orally Bioavailable Brain Penetrant Leads.
J.Med.Chem., 58, 2015
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
