8GD5
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
8ORI
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (orthorhombic) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6YN2
 
 | |
1ZZZ
 
 | | Trypsin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, CALCIUM ION, TRYPSIN | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-06-02 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
8GD3
 
 | | Crystal Structure of HIV-1 LM/HT Clade A/E CRF01 GP120 Core in Complex with DL-I-101 | | Descriptor: | (3S,5R)-N-(4-chloro-3-fluorophenyl)-1-(4-glycylpiperazine-1-carbonyl)-5-(hydroxymethyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Nguyen, D.N, Pazgier, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Piperidine CD4-Mimetic Compounds Expose Vulnerable Env Epitopes Sensitizing HIV-1-Infected Cells to ADCC.
Viruses, 15, 2023
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
6GP3
 
 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, inactive form | | Descriptor: | CALCIUM ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Srinivas, V, Lebrette, H, Lundin, D, Kutin, Y, Sahlin, M, Lerche, M, Enrich, J, Branca, R.M.M, Cox, N, Sjoberg, B.M, Hogbom, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens.
Nature, 563, 2018
|
|
6GQP
 
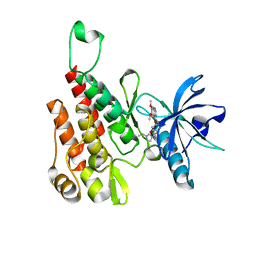 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 23) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(1-ethylpyrazol-4-yl)ethanamide | | Authors: | Hardy, C.J, Schimpl, M, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7BWM
 
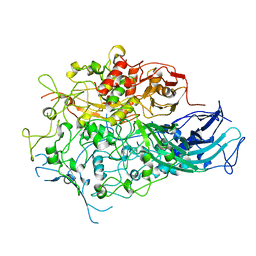 | |
6DHE
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.05 Angstrom resolution | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1NFF
 
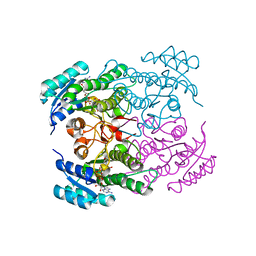 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6H6O
 
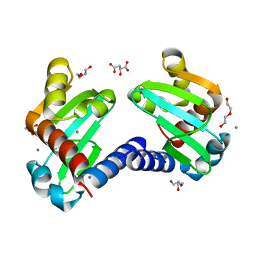 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6S48
 
 | | AvaII RESTRICTION ENDONUCLEASE IN COMPLEX WITH PARTIALLY CLEAVED dsDNA | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, DNA (5'-D(*GP*AP*TP*G)-3'), ... | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-26 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
4C7Q
 
 | |
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6GJS
 
 | | Human NBD1 of CFTR in complex with nanobodies D12 and T4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GK4
 
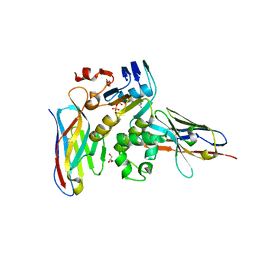 | | Human NBD1 of CFTR in complex with nanobodies D12 and T8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
1NA8
 
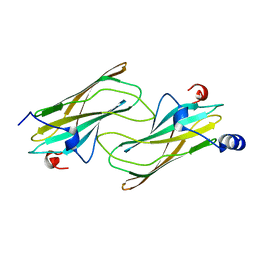 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
8DWO
 
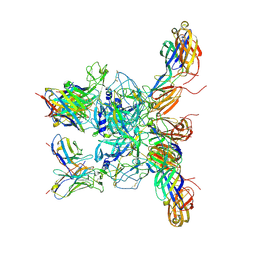 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
6QGS
 
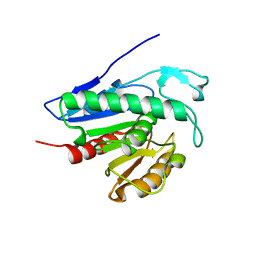 | | Crystal structure of APT1 bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
6GQQ
 
 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 35) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
