8EP2
 
 | |
8EMB
 
 | | X-ray crystal structure of Thermosynechococcus elongatus Si3 domain of RNA polymerase RpoC2 subunit | | Descriptor: | DNA-directed RNA polymerase subunit beta' | | Authors: | Murakami, K.S, Imashimizu, M, Qayyum, M.Z, Vishwakarma, R.K, Yuzenkova, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8EP9
 
 | | The capsid structure of Human Parvovirus 4 | | Descriptor: | Human Parvovirus 4 | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Capsid Structure of Aleutian Mink Disease Virus and Human Parvovirus 4: New Faces in the Parvovirus Family Portrait.
Viruses, 14, 2022
|
|
1KP9
 
 | | Crystal structure of mycolic acid cyclopropane synthase CmaA1, apo-form | | Descriptor: | ACETIC ACID, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1 | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORL
 
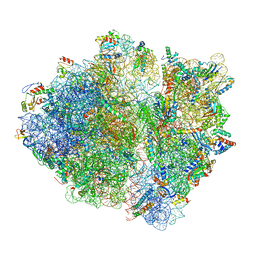 | | RF1 pre-accommodated 70S complex at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6EXK
 
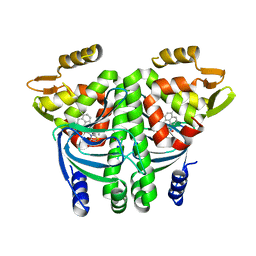 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - unfolded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
8EXP
 
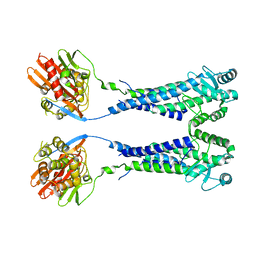 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
6ZLE
 
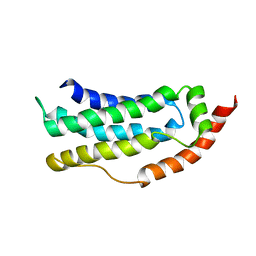 | |
5FTQ
 
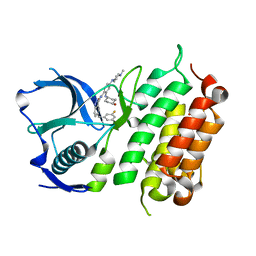 | | Crystal structure of the ALK kinase domain in complex with Cmpd 17 | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, GLYCEROL, N-[5-(3,5-DIFLUOROBENZYL)-1H-INDAZOL-3-YL]-2-[(4-HYDROXYCYCLOHEXYL)AMINO]-4-(4-METHYLPIPERAZIN-1-YL) BENZAMIDE | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
7PL9
 
 | |
8CC3
 
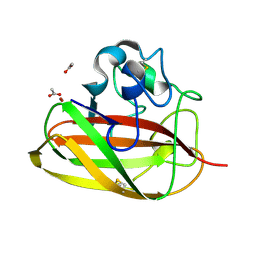 | | Vibrio cholerae GbpA (LPMO domain) | | Descriptor: | ACETATE ION, COPPER (II) ION, GlcNAc-binding protein A, ... | | Authors: | Montserrat-Canals, M, Sorensen, H.V, Cordara, G, Krengel, U. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.128 Å) | | Cite: | Perdeuterated GbpA Enables Neutron Scattering Experiments of a Lytic Polysaccharide Monooxygenase.
Acs Omega, 8, 2023
|
|
8EXR
 
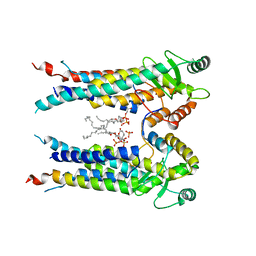 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
6ZMI
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMO
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S-eEF1a ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
5JU6
 
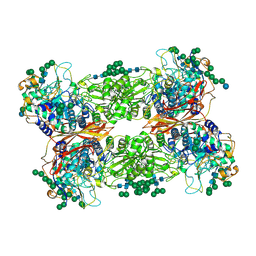 | | Structural and Functional Studies of Glycoside Hydrolase Family 3 beta-Glucosidase Cel3A from the Moderately Thermophilic Fungus Rasamsonia emersonii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Gudmundsson, M, Sandgren, M, Karkehabadi, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional studies of the glycoside hydrolase family 3 beta-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1L6J
 
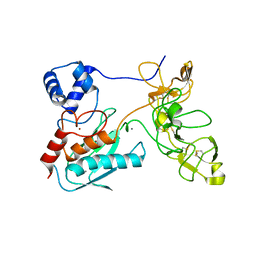 | | Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B). | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, ZINC ION | | Authors: | Elkins, P.A, Ho, Y.S, Smith, W.W, Janson, C.A, D'Alessio, K.J, McQueney, M.S, Cummings, M.D, Romanic, A.M. | | Deposit date: | 2002-03-11 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8EXQ
 
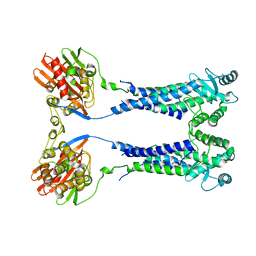 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
5OSQ
 
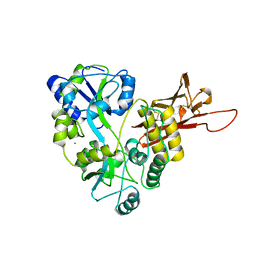 | | ZP-N domain of mammalian sperm receptor ZP3 (crystal form II, processed in P21221) | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Zona pellucida sperm-binding protein 3, TRIETHYLENE GLYCOL, ... | | Authors: | Jovine, L, Monne, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats
Nature, 456, 2008
|
|
8CC5
 
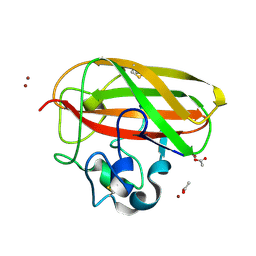 | | Vibrio cholerae GbpA (LPMO domain) | | Descriptor: | ACETATE ION, COPPER (II) ION, GlcNAc-binding protein A, ... | | Authors: | Montserrat-Canals, M, Sorensen, H.V, Cordara, G, Krengel, U. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Perdeuterated GbpA Enables Neutron Scattering Experiments of a Lytic Polysaccharide Monooxygenase.
Acs Omega, 8, 2023
|
|
8C54
 
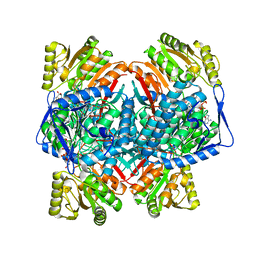 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
5C2V
 
 | | Kuenenia stuttgartiensis Hydrazine Synthase | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Dietl, A, Ferousi, C, Maalcke, W.J, Menzel, A, de Vries, S, Keltjens, J.T, Jetten, M.S.M, Kartal, B, Barends, T.R.M. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The inner workings of the hydrazine synthase multiprotein complex.
Nature, 527, 2015
|
|
6X2K
 
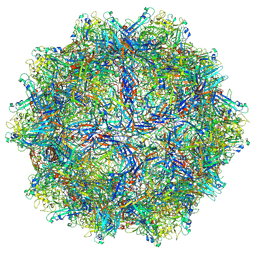 | |
8C0W
 
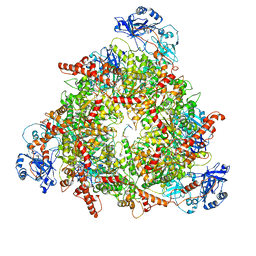 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
6WJ5
 
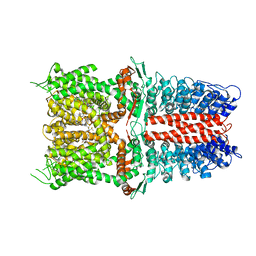 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
