3CYF
 
 | | Crystal Structure of E18N DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3CXA
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Balaji, K, Sharma, P, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution
To be Published
|
|
7EFD
 
 | | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose)
To Be Published
|
|
7X2Y
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
3CZ0
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3CZ9
 
 | | Crystal Structure of E18L DJ-1 | | Descriptor: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3D1P
 
 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
7EFC
 
 | | 1.70 A cryo-EM structure of streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | 1.70 A cryo-EM structure of streptavidin using all frames (corresponding to 70 e/A^2 total dose)
To Be Published
|
|
6IM1
 
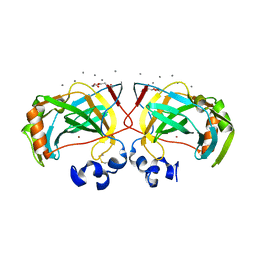 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), TETRAETHYLENE GLYCOL, ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
7E4L
 
 | |
3DTR
 
 | |
3DV3
 
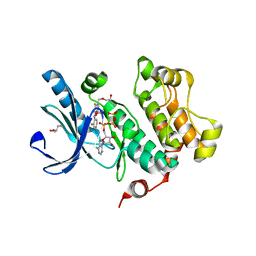 | | MEK1 with PF-04622664 Bound | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(2-hydroxyethoxy)methyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Kazmirski, S.L, Kothe, M, Ding, Y.-H. | | Deposit date: | 2008-07-18 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3DXS
 
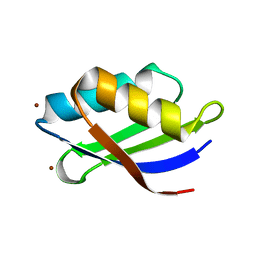 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
3DWN
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant A77E/S100R | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2008-07-22 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall.
Nature, 458, 2009
|
|
3DY3
 
 | | Crystal structure of yeast 20S proteasome in complex with the epimer form of spirolactacystin | | Descriptor: | (3R,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxo-D-proline, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Balskus, E, Jacobsen, E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural analysis of spiro beta-lactone proteasome inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3DR5
 
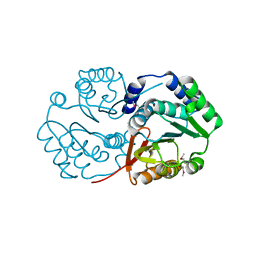 | | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium target CgR117. | | Descriptor: | Putative O-Methyltransferase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum.
To be Published
|
|
7W4K
 
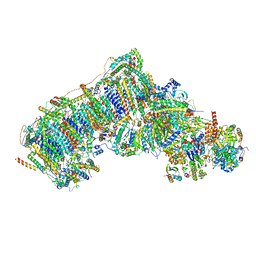 | | Deactive state CI from Q1-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4J
 
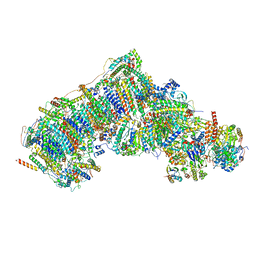 | | Deactive state CI from Q1-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3DVI
 
 | |
7WFS
 
 | |
3DWU
 
 | |
6K65
 
 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7EG2
 
 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
