8JUG
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | Descriptor: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Marsh, M.E, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8JLV
 
 | |
1DTZ
 
 | | STRUCTURE OF CAMEL APO-LACTOFERRIN DEMONSTRATES ITS DUAL ROLE IN SEQUESTERING AND TRANSPORTING FERRIC IONS SIMULTANEOUSLY:CRYSTAL STRUCTURE OF CAMEL APO-LACTOFERRIN AT 2.6A RESOLUTION. | | Descriptor: | APO LACTOFERRIN | | Authors: | Khan, J.A, Kumar, P, Paramasivam, M, Srinivasan, A, Yadav, R.S, Sahani, M.S, Singh, T.P. | | Deposit date: | 2000-01-13 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Camel lactoferrin, a transferrin-cum-lactoferrin: crystal structure of camel apolactoferrin at 2.6 A resolution and structural basis of its dual role.
J.Mol.Biol., 309, 2001
|
|
1FQV
 
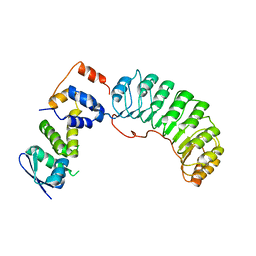 | | Insights into scf ubiquitin ligases from the structure of the skp1-skp2 complex | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-06 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
1FS2
 
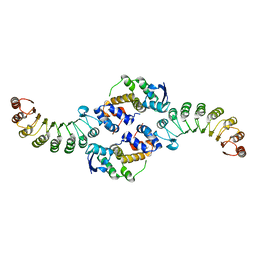 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
3VHK
 
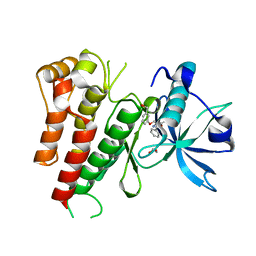 | | Crystal structure of the VEGFR2 kinase domain in complex with a back pocket binder | | Descriptor: | 1,2-ETHANEDIOL, Vascular endothelial growth factor receptor 2, {3-[(5-methyl-2-phenyl-1,3-oxazol-4-yl)methoxy]phenyl}methanol | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
1D4U
 
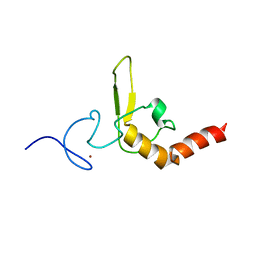 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
1FS1
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | Descriptor: | CYCLIN A/CDK2-ASSOCIATED P19, CYCLIN A/CDK2-ASSOCIATED P45 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
3VID
 
 | | Crystal structure of human VEGFR2 kinase domain with Compound A. | | Descriptor: | 4,5,6,11-tetrahydro-1H-pyrazolo[4',3':6,7]cyclohepta[1,2-b]indole, Vascular endothelial growth factor receptor 2 | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-09-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
3WC8
 
 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
3G7Y
 
 | | Crystal structure of oxidized Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7WTQ
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (without Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7XK5
 
 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 3 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7WTR
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-3 | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTP
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTS
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State UTP14 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTX
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTT
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (with CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7XK6
 
 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with aurachin D-42 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aurachin D, CALCIUM ION, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7WTZ
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTU
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (without CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7XK3
 
 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 1 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8CZZ
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
