6CJX
 
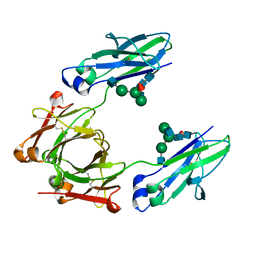 | | Crystal structure of a Fc fragment LALA mutant (L234A, L235A) of human IgG1 (crystal form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of human IgG1 | | Authors: | Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-02-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fc gamma Receptor Binding.
Structure, 2020
|
|
3WZP
 
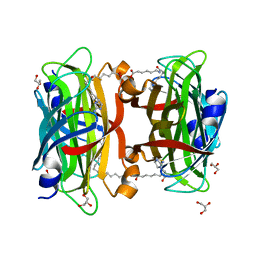 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
6CLA
 
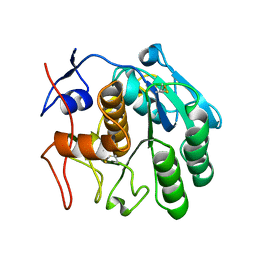 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3X0B
 
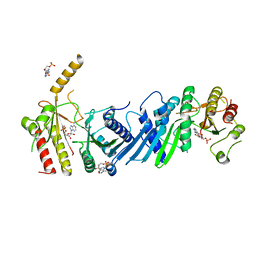 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
6CPM
 
 | | Structure of the USP15 deubiquitinase domain in complex with a third-generation inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6CLJ
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLQ
 
 | | 1.21 A MicroED structure of GSNQNNF at 2.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.21 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3X0C
 
 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X20
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 25 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, N, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5HQQ
 
 | | DNA duplex containing a ribonolactone lesion | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(RIB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Constant, J.F, Jourdan, M. | | Deposit date: | 2016-01-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of DNA Featuring Clustered 2'-Deoxyribonolactone and 8-Oxoguanine Lesions.
Biochemistry, 55, 2016
|
|
5D3L
 
 | | First bromodomain of BRD4 bound to inhibitor XD35 | | Descriptor: | 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxy-4-methylphenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3P
 
 | | First bromodomain of BRD4 bound to inhibitor XD41 | | Descriptor: | 1-[(4-acetyl-3-ethyl-5-methyl-1H-pyrrol-2-yl)carbonyl]-N-methyl-1H-indole-6-sulfonamide, Bromodomain-containing protein 4, NICKEL (II) ION | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
6CBA
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and canavanine | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, L-CANAVANINE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
4IGR
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist ZA302 | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
5D8Y
 
 | | 2.05A resolution structure of iron bound BfrB (L68A E81A) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
3ZQB
 
 | | PrgI-SipD from Salmonella typhimurium | | Descriptor: | GLYCEROL, PROTEIN PRGI, CELL INVASION PROTEIN SIPD | | Authors: | Lunelli, M, Kolbe, M. | | Deposit date: | 2011-06-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Prgi-Sipd: Insight Into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Plos Pathog., 7, 2011
|
|
5H8Y
 
 | | Crystal structure of the complex between maize sulfite reductase and ferredoxin in the form-2 crystal | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, ... | | Authors: | Kurisu, G, Nakayama, M, Hase, T. | | Deposit date: | 2015-12-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mutational studies of an electron transfer complex of maize sulfite reductase and ferredoxin.
J.Biochem., 160, 2016
|
|
3X00
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
4INS
 
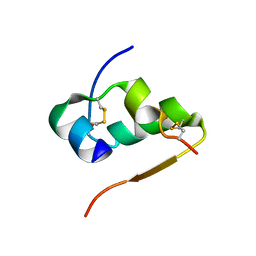 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
3WZO
 
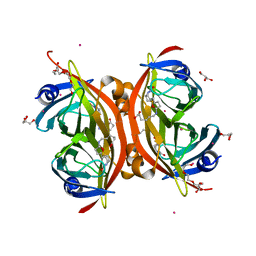 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | Descriptor: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X07
 
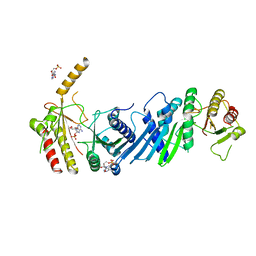 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X24
 
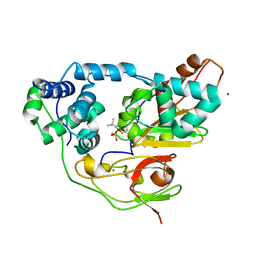 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5HQ3
 
 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
4JMW
 
 | |
5CQW
 
 | | Tetragonal Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
