5HKI
 
 | | Crystal structure of Mycobacterium tuberculosis H37Rv orotate phosphoribosyltransferase in complex with Fe(III) dicitrate | | Descriptor: | Iron(III) dicitrate, Orotate phosphoribosyltransferase | | Authors: | Donini, S, Ferraris, D.M, Bolognesi, G, Rizzi, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigations on orotate phosphoribosyltransferase from Mycobacterium tuberculosis, a key enzyme of the de novo pyrimidine biosynthesis.
Sci Rep, 7, 2017
|
|
7RBY
 
 | | Crystal structure of Nanobody nb112 and SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ilama-isolated nanobody NIH-CoV nb-112 specific to SARS-CoV-2 RBD, MAGNESIUM ION, ... | | Authors: | Chen, Y, Tolbert, W, Pazgier, M. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19.
Mabs, 14
|
|
5J01
 
 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+ and MG2+. | | Descriptor: | AMMONIUM ION, MAGNESIUM ION, group II intron lariat | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
5J0G
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P7/8 | | Descriptor: | OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
8OKI
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex bound to Spt4/5 | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
5J26
 
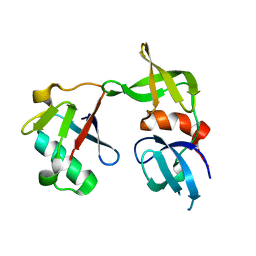 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
5HML
 
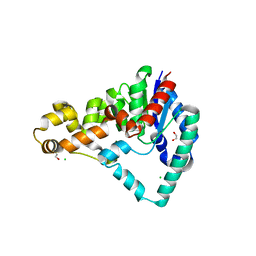 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Flemming, C.S, Feng, M, Sedelnikova, S.E, Zhang, J, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HHD
 
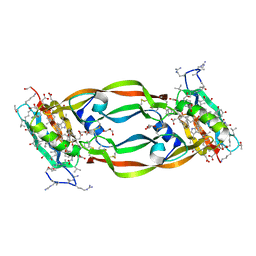 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5HN4
 
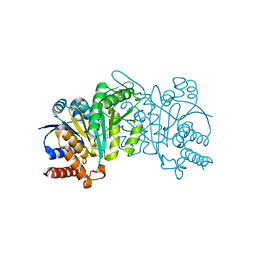 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and homoisocitrate | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, Homoisocitrate dehydrogenase, IMIDAZOLE, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
8OT7
 
 | |
5J3V
 
 | |
5EUA
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 in complex with Moxalactam | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, Beta-lactamase, SODIUM ION | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
8P2I
 
 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation with Spt4/5 | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
2ZBC
 
 | | Crystal structure of STS042, a stand-alone RAM module protein, from hyperthermophilic archaeon Sulfolobus tokodaii strain7. | | Descriptor: | 83aa long hypothetical transcriptional regulator asnC, ISOLEUCINE | | Authors: | Miyazono, K, Tsujimura, M, Kawarabayasi, Y, Tanokura, M. | | Deposit date: | 2007-10-19 | | Release date: | 2008-03-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of STS042, a stand-alone RAM module protein, from hyperthermophilic archaeon Sulfolobus tokodaii strain7
Proteins, 71, 2008
|
|
7R04
 
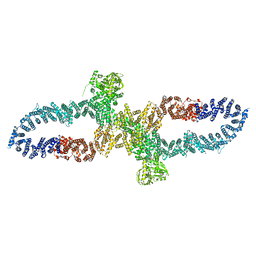 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
5J5F
 
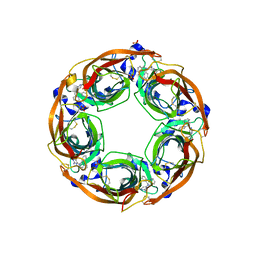 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6HEE
 
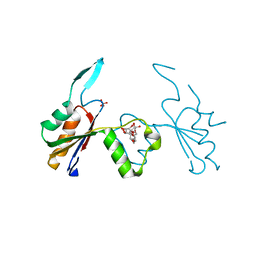 | | Crystal structure of Extracellular Domain 1 (ECD1) of FtsX from S. pneumonie in complex with undecyl-maltoside | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsX, SULFATE ION, ... | | Authors: | Martinez-Caballero, S, Alcorlo-Pages, M, Hermoso, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
Mbio, 10, 2019
|
|
5EM0
 
 | | Crystal structure of mugwort allergen Art v 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Pollen allergen Art v 4.01, SODIUM ION | | Authors: | Offermann, L.R, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
7R03
 
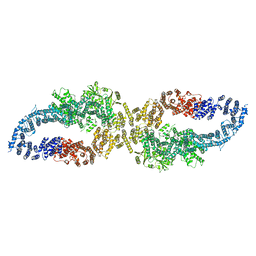 | |
8OTP
 
 | |
5EUF
 
 | | The crystal structure of a protease from Helicobacter pylori | | Descriptor: | GLYCEROL, Protease, ZINC ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a protease from Helicobacter pylori
To Be Published
|
|
7R09
 
 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ and Cyclohexylamine | | Descriptor: | Amine Dehydrogenase, CYCLOHEXYLAMMONIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
8OUB
 
 | |
6YWA
 
 | |
3UU8
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
