6I5R
 
 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | Sugar ABC transporter substrate-binding protein, BlMnBP1, ZINC ION, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
1IUH
 
 | | Crystal structure of TT0787 of thermus thermophilus HB8 | | Descriptor: | 2'-5' RNA Ligase | | Authors: | Kato, M, Sakai, H, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the 2'-5' RNA Ligase from Thermus thermophilus HB8
J.MOL.BIOL., 329, 2003
|
|
1SRO
 
 | | S1 RNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PNPASE | | Authors: | Bycroft, M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold.
Cell(Cambridge,Mass.), 88, 1997
|
|
8BFW
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-27 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
7D9X
 
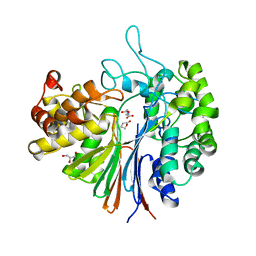 | | Highly active mutant W525D of Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens | | Descriptor: | GAMMA-BUTYROLACTONE, GLYCEROL, GLYCINE, ... | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9E
 
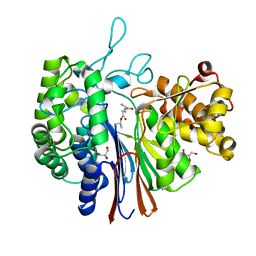 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCEROL, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
1IV4
 
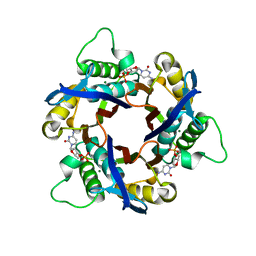 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form Substrate) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8F0W
 
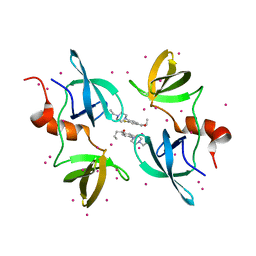 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
1IWP
 
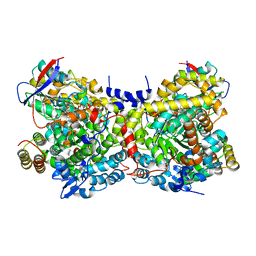 | | Glycerol Dehydratase-cyanocobalamin Complex of Klebsiella pneumoniae | | Descriptor: | COBALAMIN, Glycerol Dehydratase Alpha subunit, Glycerol Dehydratase Beta subunit, ... | | Authors: | Yamanishi, M, Yunoki, M, Tobimatsu, T, Toraya, T. | | Deposit date: | 2002-05-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol.
Eur.J.Biochem., 269, 2002
|
|
1IWC
 
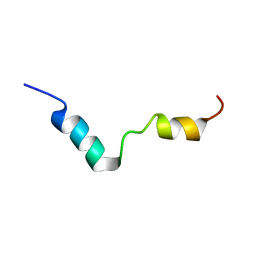 | | TFE-induded structure of the N-terminal domain of pig gastric H/K-ATPase | | Descriptor: | gastric H/K-ATPase | | Authors: | Fujitani, N, Kanagawa, M, Aizawa, T, Ohkubo, T, Kaya, S, Demura, M, Kawano, K, Taniguchi, K, Nitta, K. | | Deposit date: | 2002-05-02 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure determination and conformational change induced by tyrosine phosphorylation of the N-terminal domain of the alpha-chain of pig gastric H+/K+-ATPase
Biochem.Biophys.Res.Commun., 300, 2003
|
|
7D9W
 
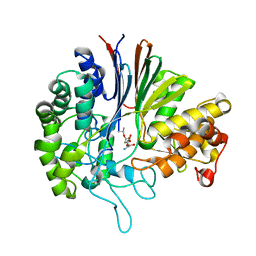 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Putthapong, P, Hayashi, J, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
1SEP
 
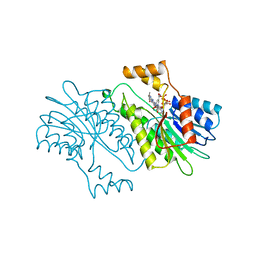 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1J0C
 
 | | ACC deaminase mutated to catalytic residue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1WIA
 
 | | Solution structure of mouse hypothetical ubiquitin-like protein BAB25500 | | Descriptor: | hypothetical ubiquitin-like protein (RIKEN cDNA 2010008E23) | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Hayashi, F, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse hypothetical ubiquitin-like protein BAB25500
To be Published
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
1J3B
 
 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4ZGC
 
 | | Crystal Structure Analysis of Kelch protein (with disulfide bond) from Plasmodium falciparum | | Descriptor: | Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-04-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kelch protein with disulfide bond from Plasmodium falciparum.
to be published
|
|
8EYO
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 with NADP bound | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, ... | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
3LQD
 
 | | Crystal structure determination of Lepus europaeus 2.8 A resolution | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Thenmozhi, M, Sathya Moothy, Pon, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure determination of Lepus europaeus 2.8 A resolution
To be published
|
|
6IC3
 
 | | AL amyloid fibril from a lambda 1 light chain | | Descriptor: | lambda 1 light chain fragment, residues 3-118 | | Authors: | Fritz, G, Faendrich, M, Schmidt, M, Radamaker, L. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-03 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis.
Nat Commun, 10, 2019
|
|
4ZGB
 
 | | Structure of untreated lipase from Thermomyces lanuginosa at 2.3 A resolution | | Descriptor: | Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-04-22 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
6I5H
 
 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5LLS
 
 | | Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3~{R})-3-azanylpiperidin-1-yl]-7-[(2-bromophenyl)methyl]-1,3-dimethyl-purine-2,6-dione, ... | | Authors: | Nar, H, Blaesse, M. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Comparative Analysis of Binding Kinetics and Thermodynamics of Dipeptidyl Peptidase-4 Inhibitors and Their Relationship to Structure.
J.Med.Chem., 59, 2016
|
|
8EYN
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 in Apo Form | | Descriptor: | CITRIC ACID, NADP-dependent malic enzyme, mitochondrial | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
