4BJQ
 
 | | Crystal structure of E. coli penicillin binding protein 3, domain V88- S165 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENICILLIN BINDING PROTEIN TRANSPEPTIDASE DOMAIN PROTEIN, SULFATE ION | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
6TH6
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
4JVH
 
 | | Structure of the star domain of quaking protein in complex with RNA | | Descriptor: | Protein quaking, RNA (5'-R(*UP*UP*CP*AP*CP*UP*AP*AP*CP*AP*A)-3'), SULFATE ION | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
4JVU
 
 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6NW1
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin V37N | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
6ONE
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A01. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
6OJX
 
 | |
8GPP
 
 | |
5O3J
 
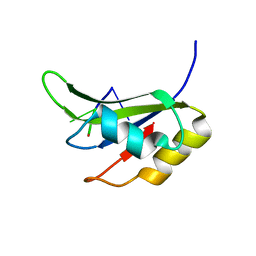 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | Descriptor: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | Authors: | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6ONH
 
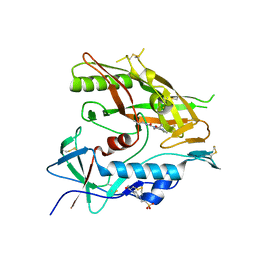 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-031-A05. | | Descriptor: | (3S)-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONV
 
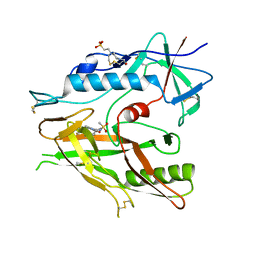 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-027-D05. | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(methylsulfonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6OOO
 
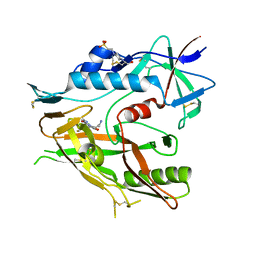 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-226. | | Descriptor: | (3S,5R)-5-amino-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Small Molecules That Sensitize HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity.
Acs Med.Chem.Lett., 11, 2020
|
|
6OOT
 
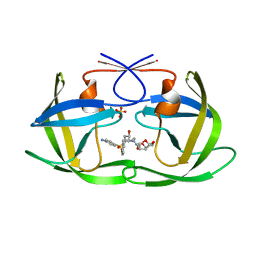 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
1B5U
 
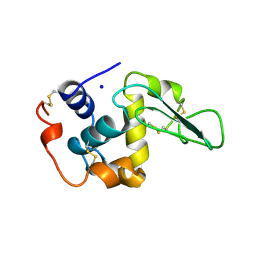 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
6TQQ
 
 | |
6MZJ
 
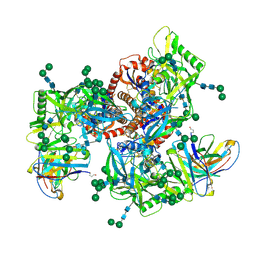 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6N09
 
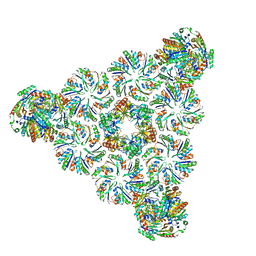 | |
8GB1
 
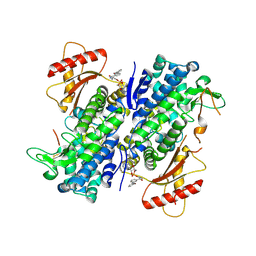 | | Crystal structure of SAMHD1 dimer bound to deoxyguanosine linked inhibitor | | Descriptor: | 5'-O-[(R)-(3-{[(1M)-3'-bromo[1,1'-biphenyl]-3-carbonyl]amino}propoxy)(hydroxy)phosphoryl]-2'-deoxyguanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
8GSY
 
 | |
2V8F
 
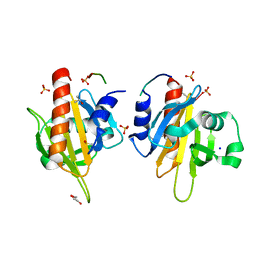 | | Mouse Profilin IIa in complex with a double repeat from the FH1 domain of mDia1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Kursula, I, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
6OKV
 
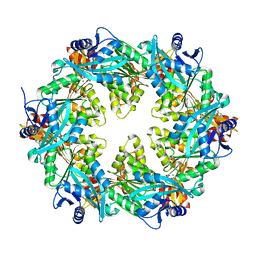 | |
6T2Q
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16, ... | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1S6D
 
 | | Structure in solution of a methionine-rich 2S Albumin protein from Sunflower Seed | | Descriptor: | Albumin 8 | | Authors: | Pantoja-Uceda, D, Bruix, M, Shewry, P.R, Santoro, J, Rico, M. | | Deposit date: | 2004-01-23 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Biochemistry, 43, 2004
|
|
