4AFB
 
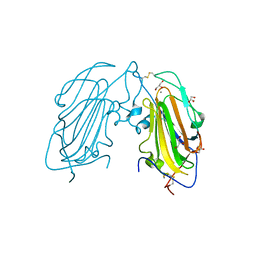 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 3 A domain (Epa1to3A) from Candida glabrata in complex with glycerol | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ASL
 
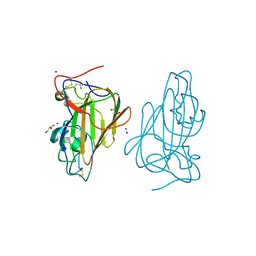 | | Structure of Epa1A in complex with the T-antigen (Gal-b1-3- GalNAc) | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B3S
 
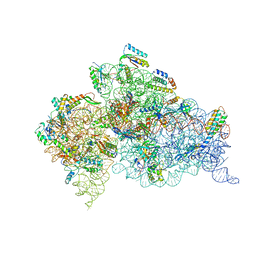 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
1SIX
 
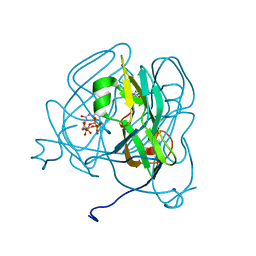 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.-S, Kim, M, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
2BHL
 
 | | X-RAY STRUCTURE OF HUMAN GLUCOSE-6-PHOSPHATE DEHYDROGENASE (DELETION VARIANT) COMPLEXED WITH GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL | | Authors: | Kotaka, M, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5AOY
 
 | | Structure of mouse Endonuclease V | | Descriptor: | ENDONUCLEASE V | | Authors: | Vik, E.S, Nawaz, M.S, Ronander, M.E, Solvoll, A.M, Strom-Andersen, P, Bjoras, M, Alseth, I, Dalhus, B. | | Deposit date: | 2015-09-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Md Simulation of Mouse Endov Reveal Wedge Motif Plasticity in This Inosine-Specific Endonuclease.
Sci.Rep., 6, 2016
|
|
5KUD
 
 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
6SVQ
 
 | | Crystal structure of human GFAT-1 G461E after UDP-GlcNAc soaking | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate-aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5AHF
 
 | | Crystal structure of Salmonella enterica HisA D7N with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
6SVM
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GalNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5AHE
 
 | | Crystal structure of Salmonella enterica HisA | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
6SVO
 
 | | Crystal structure of human GFAT-1 in complex with Glucosamine-6-Phosphate and L-Glu | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5A3G
 
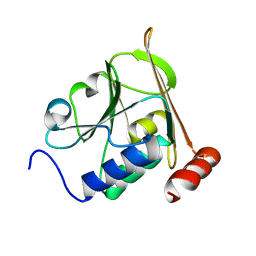 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5J9A
 
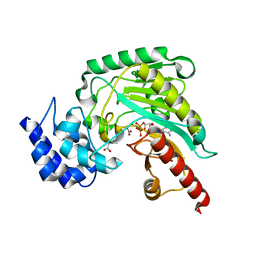 | | Ambient temperature transition state structure of arginine kinase - crystal 11/Form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
5AHI
 
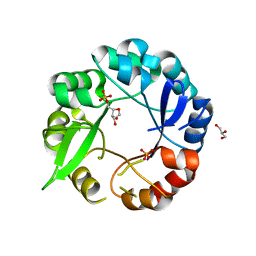 | | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanism of Hisa from Salmonella Enterica
To be Published
|
|
5J99
 
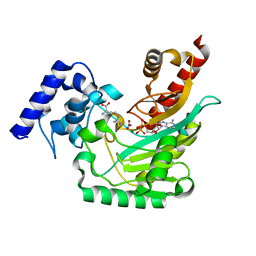 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
6SVP
 
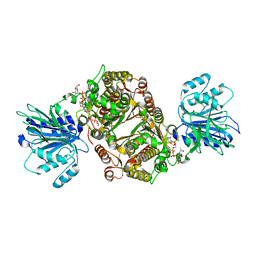 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4H
 
 | | Crystal structure of human GFAT-1 G451E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4I
 
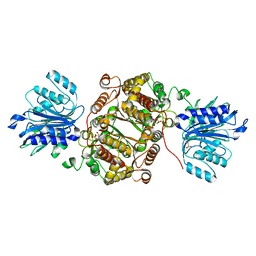 | | Crystal structure of human GFAT-1 G461E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
4P9W
 
 | | Structure of ConA/Rh3Man | | Descriptor: | 2-[2-(2-{4-[(alpha-D-mannopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Sakai, F, Weiss, M.S, Jiang, M. | | Deposit date: | 2014-04-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of ConA/Rh3Man
To Be Published
|
|
6R4J
 
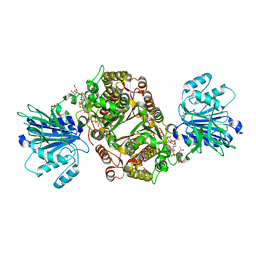 | | Crystal structure of human GFAT-1 G451E in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4E
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate and L-Glu | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5COD
 
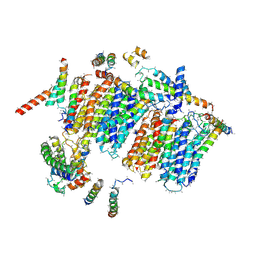 | | Bovine heart complex I membrane domain | | Descriptor: | NADH-ubiquinone oxidoreductase chain 4, NADH-ubiquinone oxidoreductase chain 5, SDAP, ... | | Authors: | Zhu, J, Hirst, J, King, M.S, Yu, M, Leslie, A.G.W, Klipcan, L. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.74 Å) | | Cite: | Structure of subcomplex I beta of mammalian respiratory complex I leads to new supernumerary subunit assignments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KUC
 
 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5A5W
 
 | | Crystal structure of Salmonella enterica HisA D7N D176A with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
