7Z4N
 
 | | Plasmodium falciparum pyruvate kinase complexed with pyruvate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z4Q
 
 | | Plasmodium falciparum pyruvate kinase mutant - C49A | | Descriptor: | MAGNESIUM ION, Pyruvate kinase | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7YZX
 
 | | ScpA from Streptococcus pyogenes, D783A mutant. | | Descriptor: | C5a peptidase, CALCIUM ION, MALONIC ACID, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Jain, M. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray diffraction data for the C5a-peptidase mutant with modified activity and specificity.
Data Brief, 46, 2023
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7YL9
 
 | | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin | | Descriptor: | Hemolysin | | Authors: | Mondal, A.K, Sengupta, N, Singh, M, Biswas, R, Lata, K, Lahiri, I, Dutta, S, Chattopadhyay, K. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin
J.Biol.Chem.
|
|
7Z1N
 
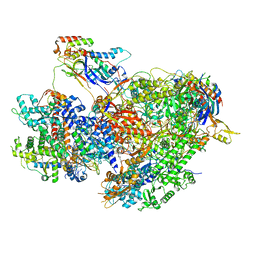 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
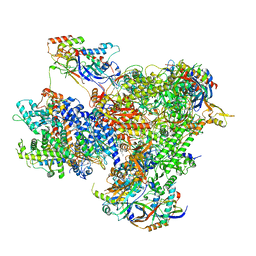 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7ZC2
 
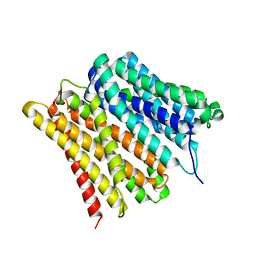 | | Dipeptide and tripeptide Permease C (DtpC) | | Descriptor: | Amino acid/peptide transporter | | Authors: | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
7Z3J
 
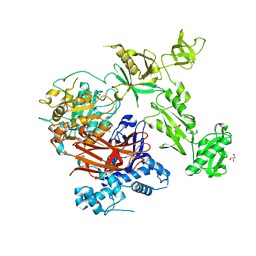 | | Structure of crystallisable rat Phospholipase C gamma 1 in complex with inositol 1,4,5-trisphosphate | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Pinotsis, N, Bunney, T.D, Katan, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the membrane interactions of phospholipase C gamma reveals key features of the active enzyme.
Sci Adv, 8, 2022
|
|
7ZEP
 
 | | Human SLFN11 E209A dimer | | Descriptor: | Schlafen family member 11, ZINC ION | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
7N12
 
 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7N11
 
 | |
7ZCE
 
 | |
7Z39
 
 | | Structure of Belumosudil bound to CK2alpha | | Descriptor: | 2-[3-[4-(1~{H}-indazol-5-ylamino)quinazolin-2-yl]phenoxy]-~{N}-propan-2-yl-ethanamide, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the Rho-associated coiled-coil kinase 2 inhibitor belumosudil bound to CK2 alpha.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ZCF
 
 | |
7Z20
 
 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7Z6N
 
 | | Crystal structure of Zn2+-transporter BbZIP in a metal-stripped state | | Descriptor: | Putative membrane protein, SULFATE ION | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
7Z6M
 
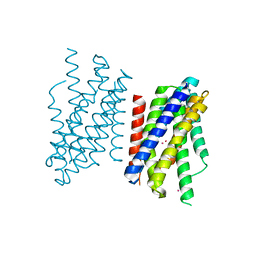 | | Crystal structure of Zn2+-transporter BbZIP in a cadmium bound state | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | Deposit date: | 2022-03-13 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
221L
 
 | |
7ZC8
 
 | |
7Z04
 
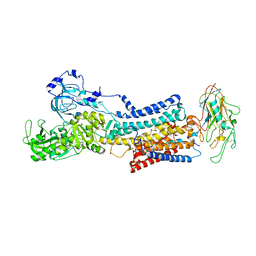 | | 10 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7YZR
 
 | | 50 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (6.92 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
8F4F
 
 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4H
 
 | | RT XFEL structure of Photosystem II 1200 microseconds after the third illumination at 2.10 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8EZ5
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
