3PH7
 
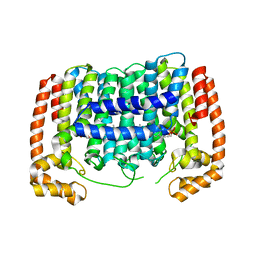 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | Farnesyl pyrophosphate synthase, GERANYLGERANYL DIPHOSPHATE | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
3D73
 
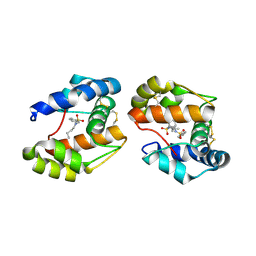 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, at pH 7.0 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
4QV0
 
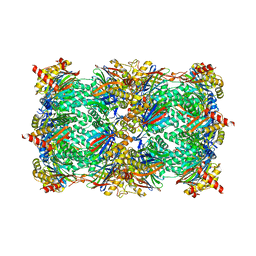 | | yCP beta5-A49T-A50V-double mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QVY
 
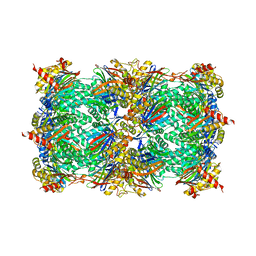 | | yCP beta5-A49T-mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWR
 
 | | yCP beta5-C52F mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QZA
 
 | | Mouse Tdt in complex with a DSB substrate, C-C base pair | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
8SGE
 
 | | KLHDC2 Kelch Domain with ligand KDRLKZ-1 | | Descriptor: | GLYCEROL, Kelch domain-containing protein 2, [(5P)-5-{3-[(2R)-butan-2-yl]-7-[(2-methoxyethoxy)carbonyl]-2-oxo-5,6,7,8-tetrahydro-1,7-naphthyridin-1(2H)-yl}-2-oxopyridin-1(2H)-yl]acetic acid | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SGF
 
 | | KLHDC2 Kelch Domain with KLHDC2 c-terminal peptide bound | | Descriptor: | GLYCEROL, HIS-SER-VAL-ASN-GLN-ARG-PHE-GLY-SER-ASN-ASN-THR-SER-GLY-SER, Kelch domain-containing protein 2 | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SH2
 
 | | KLHDC2 in complex with EloB and EloC | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 2 | | Authors: | Digianantonio, K.M, Bekes, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4QZD
 
 | | Mouse Tdt, F405A mutant, in complex with a DSB substrate, C-C base pair | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZH
 
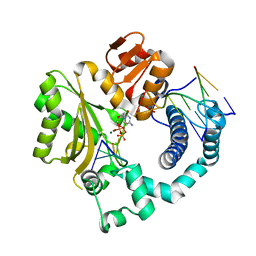 | | Mouse Tdt, F401A mutant, in complex with a DSB substrate, C-T base pair | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*T)-3', ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4R00
 
 | | yCP beta5-C52F mutant in complex with Omuralide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Omuralide, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
8T2B
 
 | |
4R3U
 
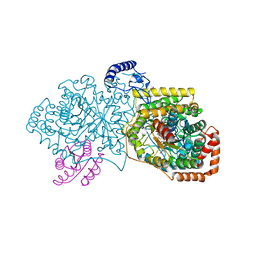 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
8T2O
 
 | |
8T2A
 
 | |
8T29
 
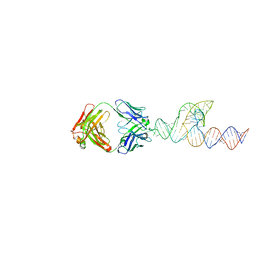 | | Crystal structure of SCV PTE RNA in complex with Fab BL3-6 | | Descriptor: | BL3-6 Fab heavy chain, BL3-6 Fab light chain, RNA (90-MER) | | Authors: | Ojha, M, Koirala, D. | | Deposit date: | 2023-06-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structure of saguaro cactus virus 3' translational enhancer mimics 5' cap for eIF4E binding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4R9J
 
 | | L-ficolin complexed to glucosamine-6-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ACETATE ION, ... | | Authors: | Laffly, E, Lacroix, M, Martin, L, Vassal-Stermann, E, Thielens, N, Gaboriaud, C. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human ficolin-2 recognition versatility extended: An update on the binding of ficolin-2 to sulfated/phosphated carbohydrates.
Febs Lett., 588, 2014
|
|
8T12
 
 | |
4RAC
 
 | | Aza-acyclic nucleoside phosphonates containing a second phosphonate group as inhibitors of the human, Plasmodium falciparum and vivax 6-oxopurine phosphoribosyltransferases and their pro-drugs as antimalarial agents | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [(2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl][(E)-2-phosphonoethenyl]amino}ethoxy)methyl]phosphonic acid | | Authors: | Keough, D.T, Hockova, D, Janeba, Z, Wang, T.-H, Naesens, L, Edstein, M.D, Chavchich, M, Guddat, L.W. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aza-acyclic Nucleoside Phosphonates Containing a Second Phosphonate Group As Inhibitors of the Human, Plasmodium falciparum and vivax 6-Oxopurine Phosphoribosyltransferases and Their Prodrugs As Antimalarial Agents.
J.Med.Chem., 58, 2015
|
|
8T13
 
 | |
4R7R
 
 | | Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | GLYCEROL, Putative lipoprotein | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To be Published
|
|
4RE3
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
8SPV
 
 | | PS3 F1 Rotorless, no ATP | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
