3DCQ
 
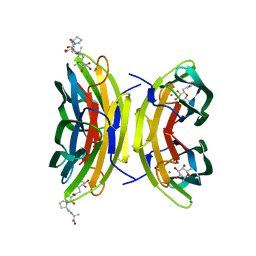 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
4U4V
 
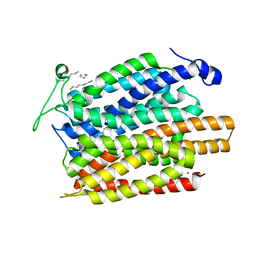 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | Descriptor: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4Y
 
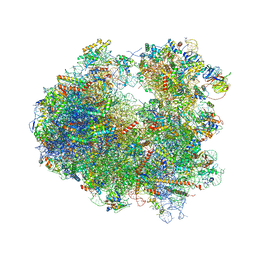 | | Crystal structure of Pactamycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4U56
 
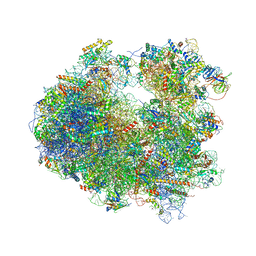 | | Crystal structure of Blasticidin S bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4TVC
 
 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with gluco-oligosaccharides | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4P4P
 
 | | Crystal structure of Leishmania infantum polymerase beta: Nick complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2973 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
4TWF
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromomemantine | | Descriptor: | Bromomemantine, Cys-loop ligand-gated ion channel | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
4P6R
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with tyrosine in the active site | | Descriptor: | TYROSINE, Tyrosinase, ZINC ION | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
4TVD
 
 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with D-glucose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
7WU8
 
 | |
3APM
 
 | | Crystal structure of the human SNP PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3APV
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
4U52
 
 | | Crystal structure of Nagilactone C bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
3AVO
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with Pantothenate | | Descriptor: | CITRATE ANION, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Chetnani, B, Kumar, P, Abhinav, K.V, Chhibber, M, Surolia, A, Vijayan, M. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Location and conformation of pantothenate and its derivatives in Mycobacterium tuberculosis pantothenate kinase: insights into enzyme action
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AWE
 
 | | Crystal structure of Pten-like domain of Ci-VSP (248-576) | | Descriptor: | ACETIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Matsuda, M, Sakata, S, Takeshita, K, Suzuki, M, Yamashita, E, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-03-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity
J.Biol.Chem., 286, 2011
|
|
3AU3
 
 | | Crystal structure of armadillo repeat domain of APC | | Descriptor: | Adenomatous polyposis coli protein | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
4UAN
 
 | |
3AUJ
 
 | | Structure of diol dehydratase complexed with glycerol | | Descriptor: | CALCIUM ION, COBALAMIN, Diol dehydrase alpha subunit, ... | | Authors: | Yamanishi, M, Kinoshita, K, Fukuoka, M, Shibata, T, Tobimatsu, T, Toraya, T. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols
Febs J., 279, 2012
|
|
4UBK
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.40 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
4TWM
 
 | | Crystal structure of dioscorin from Dioscorea japonica | | Descriptor: | Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
4TX0
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-(2-methoxyethoxy)-3-(2-methoxyethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-(2-methoxyethoxy)-3-(2-methoxyethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bischoff, M, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Stereoselective Construction of the 5-Hydroxy Diazabicyclo[4.3.1]decane-2-one Scaffold, a Privileged Motif for FK506-Binding Proteins.
Org.Lett., 16, 2014
|
|
