2A18
 
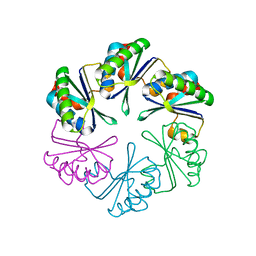 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2PYJ
 
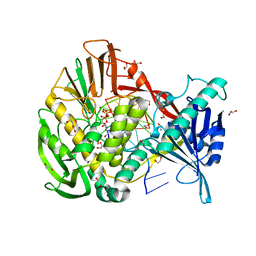 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-d(ACACGTAAGCAGTC)-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
4MOU
 
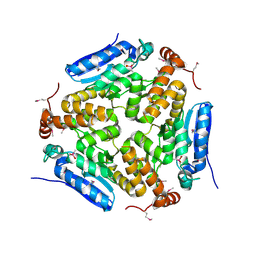 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | Descriptor: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
1HQV
 
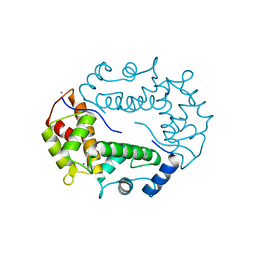 | | STRUCTURE OF APOPTOSIS-LINKED PROTEIN ALG-2 | | Descriptor: | CALCIUM ION, PROGRAMMED CELL DEATH PROTEIN 6 | | Authors: | Jia, J, Tarabykina, S, Hansen, C, Berchtold, M, Cygler, M. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apoptosis-linked protein ALG-2: insights into Ca2+-induced changes in penta-EF-hand proteins.
Structure, 9, 2001
|
|
4V1M
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
3BUA
 
 | | Crystal Structure of TRF2 TRFH domain and APOLLO peptide complex | | Descriptor: | DNA cross-link repair 1B protein, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
2A1H
 
 | | X-ray crystal structure of human mitochondrial branched chain aminotransferase (BCATm) complexed with gabapentin | | Descriptor: | ACETIC ACID, PYRIDOXAL-5'-PHOSPHATE, [1-(AMINOMETHYL)CYCLOHEXYL]ACETIC ACID, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K, Conway, M, Yennawar, N, Islam, M.M, Hutson, S.M. | | Deposit date: | 2005-06-20 | | Release date: | 2005-09-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
J.Biol.Chem., 280, 2005
|
|
7MJR
 
 | | Vip4Da2 toxin structure | | Descriptor: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | Authors: | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
2AC4
 
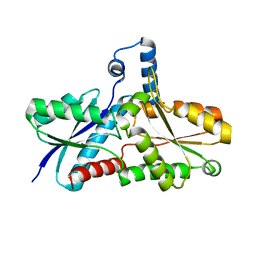 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
6ZH0
 
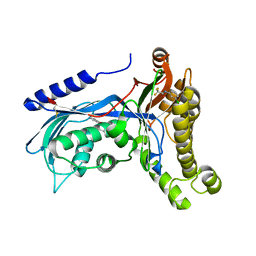 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
1HVX
 
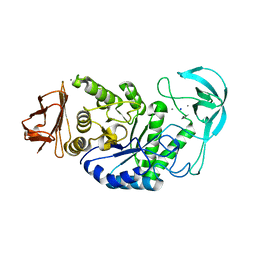 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
3C5I
 
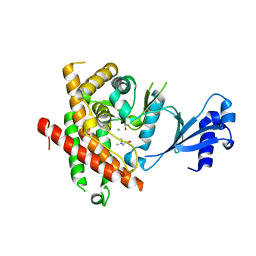 | | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520 | | Descriptor: | CALCIUM ION, CHOLINE ION, Choline kinase, ... | | Authors: | Wernimont, A.K, Hills, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
To be Published
|
|
7MPB
 
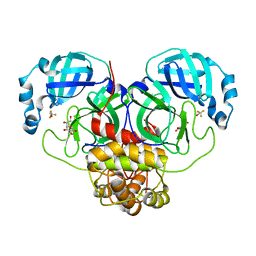 | | SARS Coronavirus-2 Main Protease 3CL-pro binding Ascorbate | | Descriptor: | 3C-like proteinase, ASCORBIC ACID, TRIFLUOROETHANOL | | Authors: | Pandey, S, Malla, T.N, Stojkovic, E.A, Schmidt, M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vitamin C inhibits SARS coronavirus-2 main protease essential for viral replication
Biorxiv, 2021
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
8DRS
 
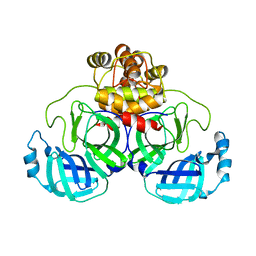 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8JG5
 
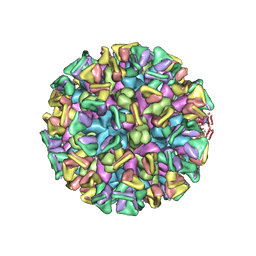 | | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | | Descriptor: | VH,SARAH, VL,SARAH, VP1 | | Authors: | Hosaka, T, Katsura, K, Kimura-Someya, T, Someya, Y, Shirouzu, M. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography revealing binding sites for human monoclonal antibodies.
J.Virol., 98, 2024
|
|
2Q5C
 
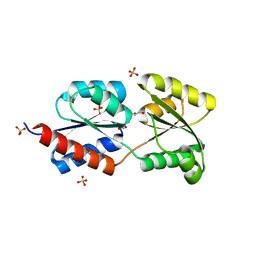 | | Crystal structure of NtrC family transcriptional regulator from Clostridium acetobutylicum | | Descriptor: | GLYCEROL, NtrC family transcriptional regulator, SULFATE ION | | Authors: | Ramagopal, U.A, Dickey, M, Toro, R, Iizuka, M, Groshong, K, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of NtrC family transcriptional regulator from Clostridium acetobutylicum.
To be Published
|
|
6EHK
 
 | | The crystal structure of CK2alpha in complex with CAM4712 and compound 37 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methyl]ethanamine, 2-hydroxy-5-methylbenzoic acid, ACETATE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
3CBJ
 
 | | Chagasin-Cathepsin B complex | | Descriptor: | Cathepsin B, Chagasin, PHOSPHATE ION | | Authors: | Redzynia, I, Bujacz, G.D, Abrahamson, M, Ljunggren, A, Jaskolski, M, Mort, J.S. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Displacement of the occluding loop by the parasite protein, chagasin, results in efficient inhibition of human cathepsin B.
J.Biol.Chem., 283, 2008
|
|
6EHU
 
 | | The crystal structure of CK2alpha in complex with compound 32 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[4-(2-ethylphenyl)-3-(trifluoromethyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1HQW
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CONCANAVALIN A WITH A TRIPEPTIDE YPY | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y, Wang, K, Cui, D, Li, M. | | Deposit date: | 2000-12-20 | | Release date: | 2003-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of concanavalin A and tripeptide
J.PROTEIN CHEM., 20, 2001
|
|
6ZTW
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
